Riboswitch aac and application thereof in preparing antibiotics
A nuclear switch and antibiotic technology, applied in the biological field, can solve the problem that erythromycin cannot be recombined, and achieve the effect of high sensitivity and universal sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1: Reporter gene carrier construction
[0053] According to the fact that aminoglycoside antibiotics are easy to act on the A site of 16S rRNA, a clustwal comparison was performed on the website of the European Biology Center, and the results showed that there are many identical bases between aac and the A site, such asfigure 1 shown.
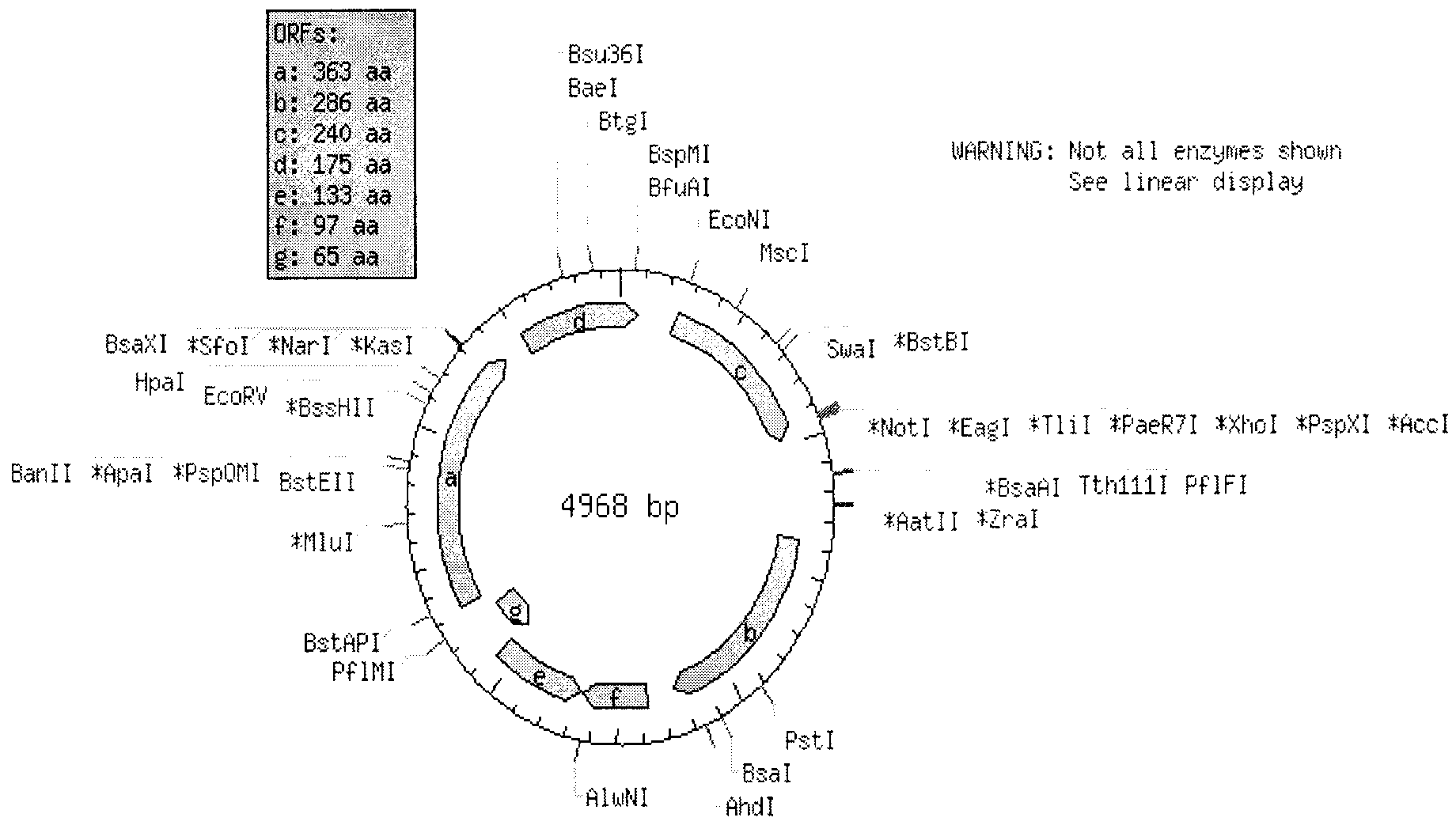
[0054] In vivo verification test, by digesting plasmid pGEX4T-3 (4968bp) at BspMI and Tth111I, inserting tac promoter (Ptac), lac operator (Olac), aac sequence, lacZα sequence, tryptophan terminator Ttrp sequence, The plasmid was named pGEX-aac-lacZα.
[0055] The resistance gene RmtB was inserted between kasI / NarI and BspMI, and the plasmid was named pGEX-RmtB-aac-lacZα. Such as Figure 2-4 shown.
Embodiment 2
[0056] Example 2: Determination of lacZ enzyme activity
[0057] Transfer 5ml of the overnight cultured JM109 containing the transformed plasmid pGEX-aac-lacZα into fresh LB liquid medium at a ratio of 1:100, add 100μg / ml ampicillin, 0.5mM IPTG, and incubate at 37°C for 1.5-2 hours; branch into sterile glass tube, add different concentrations of aminoglycoside antibiotics, and incubate for 24 hours; measure and record the absorbance of Abs600; draw 20 μl of bacterial liquid and add 80 μl of solution 1 (100 mM Na 2 HPO 4 .20mM KCl, 2mM MgSO 4 .0.8mg / mL CTAB, 0.4mg / mL sodium deoxycholate, 5.4μL / mL beta-mercaptoethanol) into 1.5ml EP tube; incubate at 30°C for 20-30min, and incubate solution 2 containing substrate ONPG under the same conditions (60mM Na 2 HPO 4 .40mM NaH 2 PO 4 .1mg / mL ONPG, 2.7μL / mLβ-mercaptoethanol); add 600μl solution 2 to each EP tube, incubate for 2h; add 700μl stop solution (1M Na 2 CO 3 ), mix well; centrifuge at 12,000 rpm for 5-10 min, and measure...
Embodiment 3
[0059] Embodiment 3: fluorescent quantitative PCR
[0060] 1. Bacterial culture
[0061] For the determination of lacZ enzyme activity, cells were collected by centrifugation after adding different concentrations of antibiotics and incubating for 24 hours.
[0062] 2. Extraction of RNA
[0063] After adding 1ml TRIzol to the sample, place it at room temperature for 5 minutes to fully lyse the sample; if the next step is not performed, the sample can be stored at -70°C for a long time; add 200 μl chloroform, vibrate vigorously and mix well, then place it at room temperature for 3-5 minutes to allow it to separate naturally. phase; centrifuge at 12,000rpm at 4°C for 10-15min. Transfer the aqueous phase (usually 550 μl) to a new tube; add an equal volume of ice-cold isopropanol to the supernatant, and place it at room temperature for 10-20 minutes; centrifuge at 12,000 rpm at 4°C for 10 minutes, discard the supernatant, and precipitate the RNA at the bottom of the tube ;Add 1m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com