Carbohydrate binding module and use thereof
一种糖类结合模块、聚糖结构的技术,应用在抑制HIV活性领域,能够解决未被实现、活性衰减、棘手安全问题等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
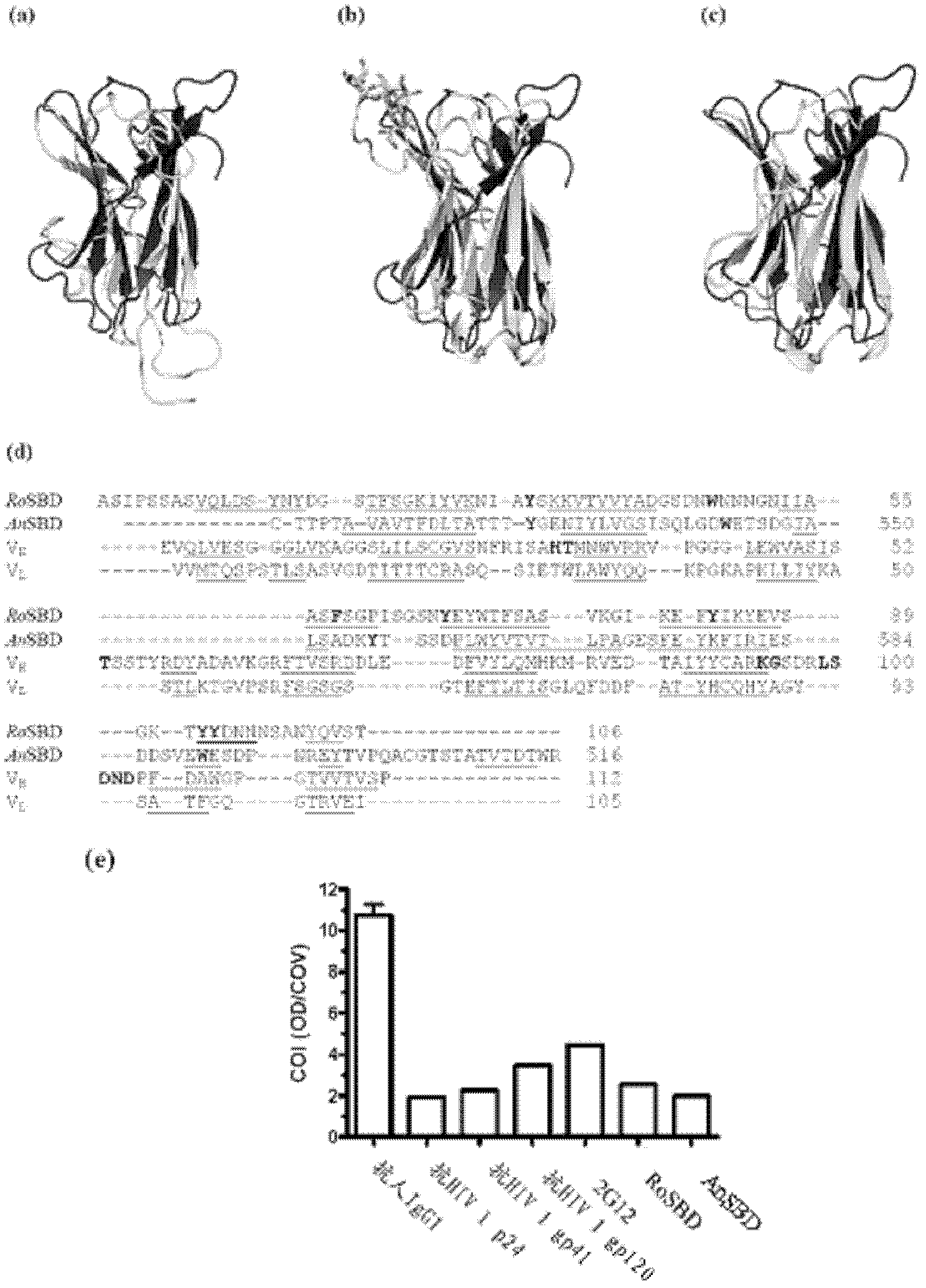
[0033] Structural relationship between SBD and 2G12
[0034] Structure-Based Multiple Sequence Alignment
[0035] Secondary structure prediction using Jpred server (server) (Cuff, J.A., Clamp, M.E., Siddiqui, A.S., Finlay, M. & Barton, GJ. JPred: a consensus secondary structure prediction server. Bioinformatics 14, 892-893 (1998)) and the Network Protein Sequence Analysis (NPSA) server. Jpred and NPSA servers provide consistent results from six (NNSSP, DSC, PREDATOR, MULPRED, ZPRED, and PHD) and twelve (SOPM, SOPMA, HNN, MLRC, DPM, DSC, GORI, GORII, GORIV, PHD , PREDATOR and SIMPA96) different methods.
[0036] result
[0037] Detection of structure-function relationships between RoSBD or AnSBD and other protein domains was performed by using the DALI database in silico (Holm, L., Kaariainen, S., Rosenstrom, P. & Schenkel, A. Searching protein structure databases with DaliLite v. 3. Bioinformatics 24, 2780-2781 (2008)). In total, 555 and 535 structurally related proteins ...
Embodiment 2
[0040] Functional relationship between SBD and 2G12
[0041] Microbes and Plasmids
[0042] Escherichia coli Top10F' (Invitrogen) was used for plasmid treatment, and E. coli BL21-Gold (DE3) (Invitrogen) was used for protein expression. Vectors pET-23a(+) and pET-15b (Novagen), both containing T7 primers (promoter), were used for expression of recombinant RoSBD and AnSBD in E. coli cells, respectively. Forward primer 5' for pET23a(+)-RoSBD
[0043] -CATATGGCAAGTATTCCCTAGCAGT-3' (SEQ ID No. 1) and reverse primer 5'-CTCGAGTCATGTAGATACTTGGT-3' (SEQ ID No. 2) containing NdeI and XhoI restriction sites, respectively, and forward primer for pET15b-AnSBD 5'-CATATGAGCAAGACCAGCACCAGT-3' (SEQ ID No. 3) and reverse primer 5'-CTCGAGCTACCGCCAGGTGT-3' (SEQ ID No. 4) containing NdeI and XhoI restriction sites, respectively, were used for cloning and sequence analysis.
[0044] Transformation and expression of SBD
[0045] The pET expression vector was transformed into Escherichia coli (E....
Embodiment 3
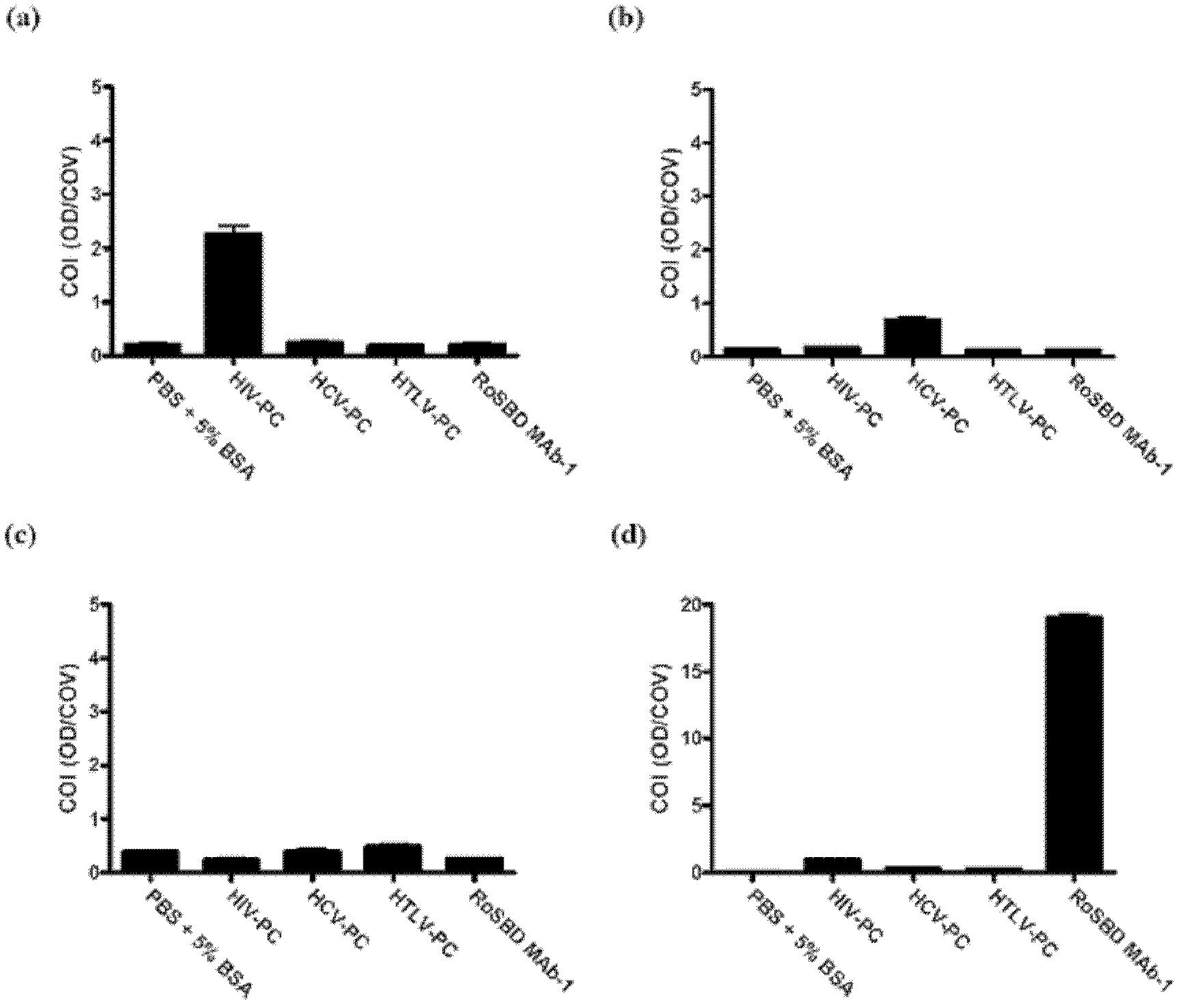
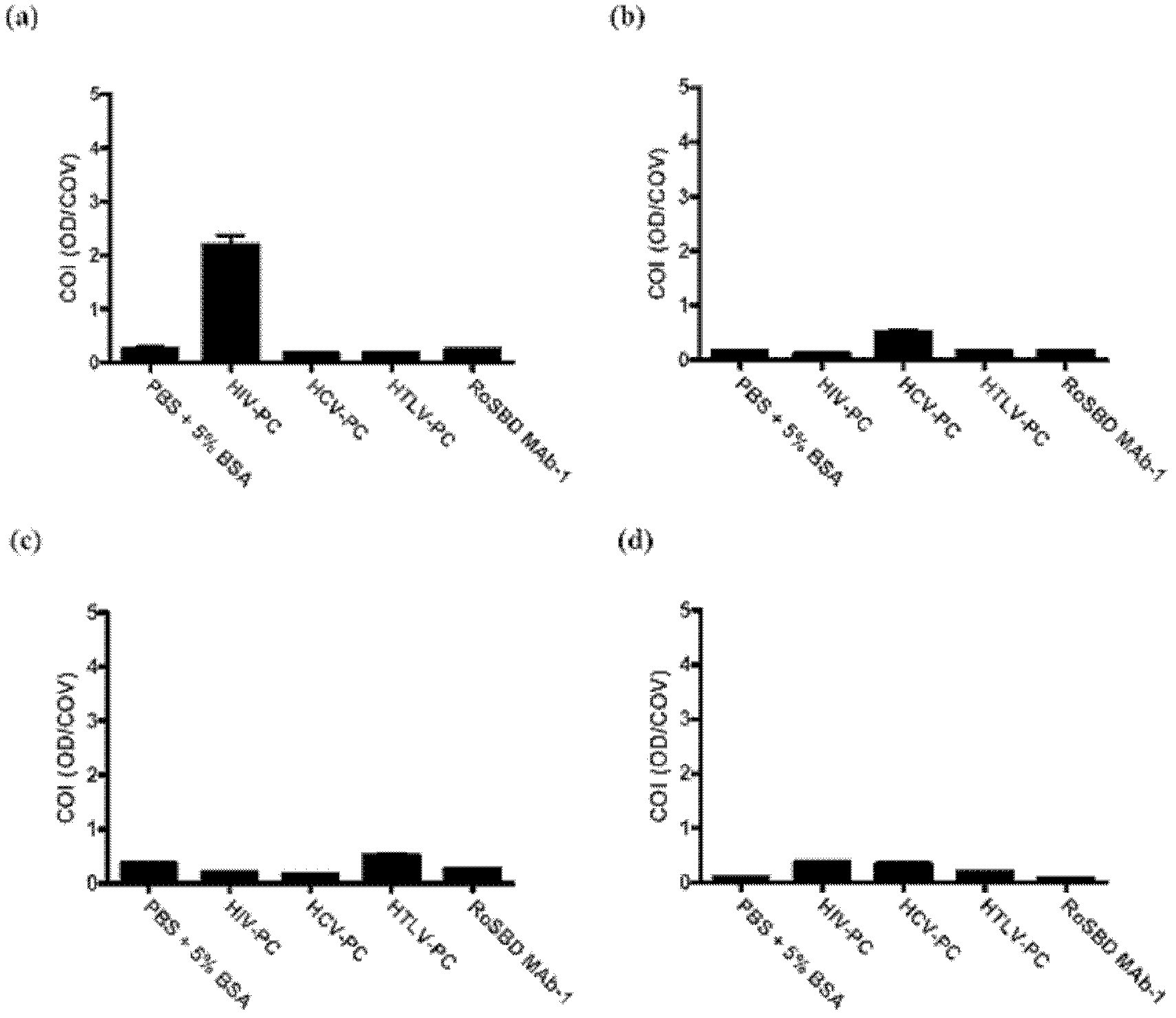
[0061] Specificity between RoSBD and AnSBD and HIV-1 glycoprotein
[0062] Competitive ELISA of HIV mAb and Glycans
[0063] The following peptides and mAbs were used in competitive ELISA experiments: (i) Human IgG 1 Secondary antibody ([2C11]; for human IgG 1 subclass-specific and not isotype-restricted; Fc-region-specific) secondary antibody, (ii) HIV-1 p24 peptide ([N29], a synthetic peptide based on N-terminal residues 1-104 of HIV-1 p24 ), (iii) HIV-1 gp120 peptide ([ED8.D4]; synthetic peptide of residues 427-448 of sequestered gp160 according to human HIV-1 BH8), (iv) HIV-1 gp41 peptide ([AG10H9], according to Synthetic peptide of residues 721-744 of HIV-1 gp160), (v) 2G12 mAb (recombinant human mAb against HIV-1 gp120), (vi) gp140 (HIV clade A, strain 92 / UG / 037). All were from Abeam, except 2G12 and gp140, which were purchased from Polymun Scientific.
[0064] For ELISA experiments, 50 μL of HIV-PC was mixed with an equal volume of 500 nM of each of (i) to (vi) abov...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com