Efficient directional seamless DNA (deoxyribonucleic acid) segment connecting method
A connection method and fragment technology, which is applied in the field of genetic engineering, can solve the problems of small number of one-time connection fragments, low degree of freedom of operation, and short length, etc., to reduce the risk of mismatch, high degree of freedom of operation, and long connection length Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
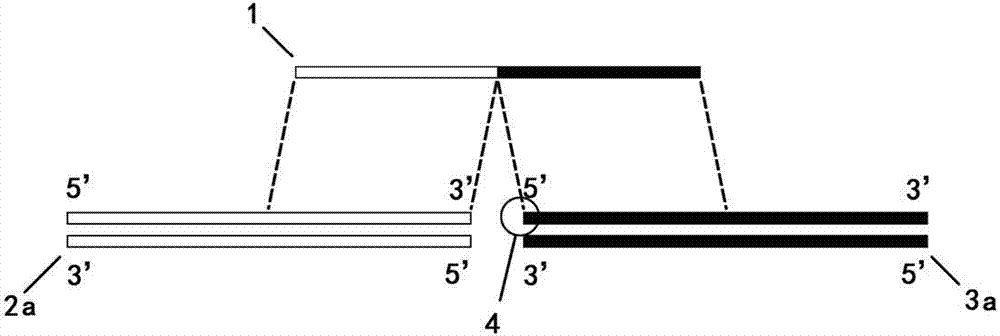
[0041] This example is directed to ligation of a blunt-ended fragment to a blunt-ended vector.
[0042] Step 1, prepare the vector 16 required for ligation and the fragment 17 to be ligated by enzyme digestion
[0043] In this example, the sequences at both ends of the vector 16 blunt end restriction site are (the arrow indicates the restriction site):
[0044] 5'-CGAATTCCTGCAGCCC ↓ GGGGGATCCACTAGTTCTAGA-3' (SEQ.ID.NO.1)
[0045] The sequences at both ends of fragment 17 to be inserted are:
[0046] 5'-GCTCTAGAATGGCAGACAATTTTTCGCTC---ATATCCCCTTTGTAAGTCATCTACTTAAGGATCCCG-3' (omitted in the middle) (SEQ.ID.NO.2)
[0047] Design and synthesize connecting primer 18, a total of two segments, corresponding to the two interfaces, its sequence is (the spaces in the sequence only indicate the parts that are complementary to the two fragments, and the actual primers are continuous without disconnection in the middle):
[0048] 18-1: 5'-CGAATTCCTGCAGCCCGCTCTAGAATGGCAGACAATTT-3' (SEQ.I...
Embodiment 2
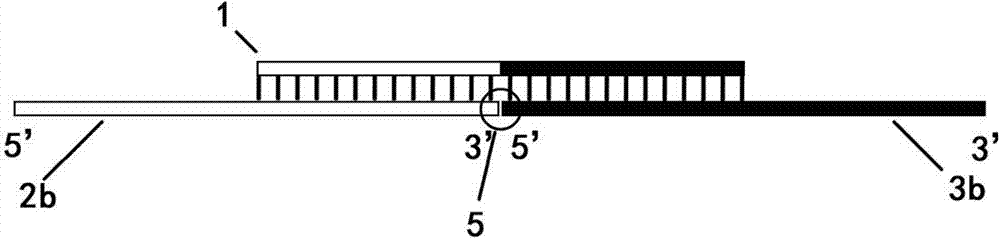
[0064] This example is directed to the ligation of a blunt-ended fragment to a cohesive-ended vector.
[0065] Step 1, prepare the vector 16 required for ligation and the fragment 17 to be ligated by enzyme digestion
[0066] In this example, the sequences at both ends of the vector 16 blunt end restriction site are (the arrow indicates the restriction site):
[0067] Xho1 cut
[0068] 5'-GGATCTATTTCCGGTGAATTCC↓TCGAGACTAGTTCTAGAGCGGC-3'
[0069] (SEQ.ID.NO.5)
[0070] 3'-CCTAGATAAAGGCCACTTAAGGAGCT↑CTGATCAAGATCTCGCCG-5'
[0071] (SEQ.ID.NO.6)
[0072] Use the klenow large fragment of E. coli DNA polymerase Ⅰ to fill in the two cohesive ends, and the sequences at both ends of the interface become:
[0073] 5'-GGATCTATTTCCGGTGAATTCCTCGA-3' (SEQ.ID.NO.7)
[0074] 5'-TCGAGACTAGTTCTAGAGCGGC-3' (SEQ.ID.NO.8)
[0075] 3'-CCTAGATAAAGGCCACTTAAGGAGCT-5' (SEQ.ID.NO.9)
[0076] 3'-AGCTCTGATCAAGATCTCGCCG-5' (SEQ.ID.NO.10)
[0077] The sequences at both...
Embodiment 3
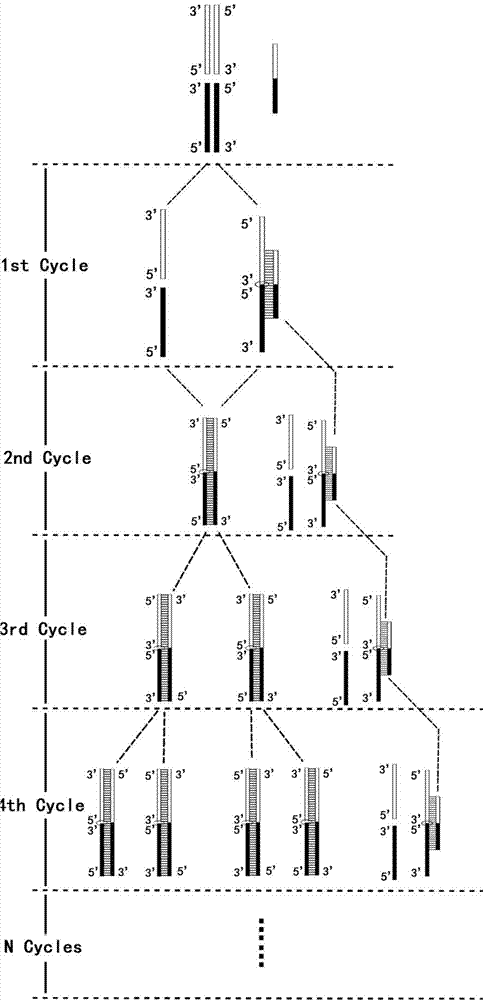
[0096] This example is aimed at the situation where 5 fragments are connected at one time to form a circular plasmid.
[0097] Step 1, prepare the vector 16 required for ligation and the fragment 17 to be ligated by enzyme digestion
[0098] In this example, the sequences at both ends of the vector 16 blunt end restriction site are (the arrow indicates the restriction site):
[0099] 5'-CGAATTCCTGCAGCCC ↓ GGGGGATCCACTAGTTCTAGA-3' (SEQ.ID.NO.14)
[0100] There are 4 pieces of 17 fragments to be inserted, and the sequences at both ends are (the middle is omitted):
[0101] 17-1: 5'-CCCAAGCTTATGGCGG---CAAAGTATAGGGGATCCCG-3' (SEQ.ID.NO.15)
[0102] 17-2: 5'-GCGCCTCCTAGTGAAAC---GTTACCCACTCCCCTCAC-3' (SEQ.ID.NO.16)
[0103] 17-3: 5'-GCATTGAGGAGATGGATGG---ATTTATGGAGCAGCTACGT-3'
[0104] (SEQ.ID.NO.17)
[0105] 17-4: 5'-GCTCTAGAATGGCAGACAATTT---AGTCATCTACTTAAGGATCCCG-3'
[0106] (SEQ.ID.NO.18)
[0107] Design and synthesize connecting primer 18, a total of 5 segment...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com