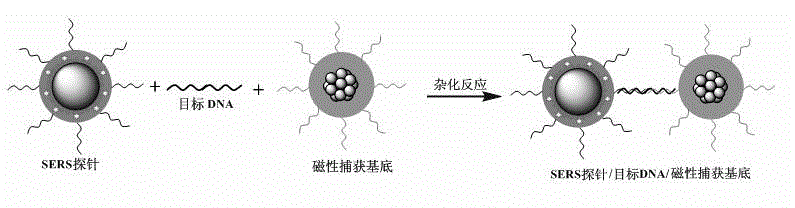
Method for high-sensitivity detection for t-DNA (transfer-deoxyribose nucleic acid) by aid of SERS (surface enhanced Raman spectroscopy) liquid chip
A sensitive detection and liquid-phase chip technology, which is applied in the field of SERS liquid-phase chips for highly sensitive detection of t-DNA, can solve the problems of dispersing the signal intensity of SERS probes, limiting the binding capacity of c-DNA, and reducing the hybridization efficiency of DNA strands, etc. Achieve the effect of improving detection sensitivity, improving capture efficiency, and increasing detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Capture DNA (c-DNA): 5’NH 2 (A 10 )TCTATAAACCTTATT (SEQ.ID.NO.1)
[0030] Target DNA (t-DNA): AGATATTTGGAATAACATGACCTGGATGCA (SEQ.ID.NO.2)
[0031] Probe DNA (p-DNA): GTACTGGACCTACGT(A 10 )NH 2 3' (SEQ. ID. NO. 3)
[0032] 1. Preparation of SERS probes
[0033] The preparation of SERS probes is completed in the following three steps:
[0034] The first step is to modify the surface of the SERS tag to amino groups:
[0035] Disperse 10 mg of the SERS tag embedded in 4-aminothiophenol (4-ABT) reported in the patent (CN102206357A) in a mixed solution of 9 g of water and 32 g of ethanol, and then add 1 g of ammonia water and 0.1 g of 3-ammonia Propyltriethoxysilane (APTES) was ultrasonically emulsified for 1 h with an ultrasonic power of 600 W to obtain SERS-labeled microspheres with surface-modified amino groups.
[0036] The second step is to modify the amino groups on the surface of the SERS label to carboxyl groups:
[0037] Add 20 mg of SERS tag with surface-mod...
Embodiment 2
[0047] Capture DNA (c-DNA): 5'NH 2 (A 10 ) AACCGAAAGTCAATA (SEQ.ID.NO.4)
[0048] Target DNA (t-DNA): TTGGCTTTCAGTTATATGGATGATGTGGTA (SEQ.ID.NO.5)
[0049] Probe DNA (p-DNA): TACCTACTACACCAT (A 10 )NH 2 3' (SEQ.ID.NO.6)
[0050] 1. The preparation of the SERS probe is the same as that described in Example 1-1. The difference is that the Raman marker molecule embedded in the SERS tag is 4-chlorothiophenol (4-CBT).
[0051] 2. The preparation of the magnetic capture substrate is the same as described in Examples 1-2.
[0052] 3. The process of detecting t-DNA by using the SERS liquid phase chip method is the same as that described in Examples 1-3.
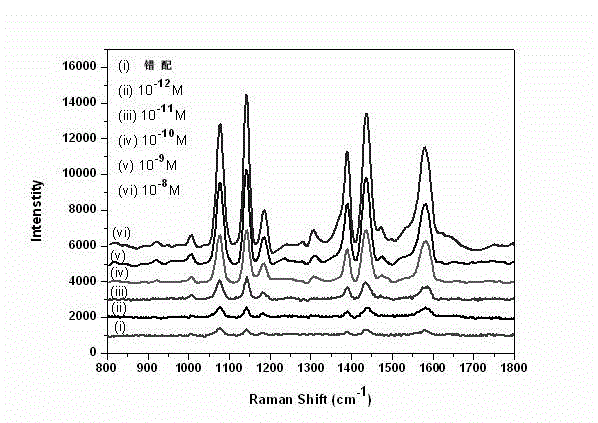
[0053] A detection limit of 10 was obtained -12 M.
Embodiment 3
[0055] Capture DNA (c-DNA): 5’NH 2 (A 10 )AATCTCAACGTACCT (SEQ.ID.NO.7)
[0056] Target DNA (t-DNA): TTAGAGTTGCATGGATTAACTCCTCTTTCT (SEQ.ID.NO.8)
[0057] Probe DNA (p-DNA): AATTGAGGAGAAAGA (A 10 )NH 2 3' (SEQ.ID.NO.9)
[0058] 1. The preparation of the SERS probe is the same as that described in Example 1-1. The difference is that the Raman marker molecule embedded in the SERS tag is 5,5'-dithiobis(2-nitrobenzoic acid) (DTNB).
[0059] 2. The preparation of the magnetic capture substrate is the same as described in Examples 1-2.
[0060] 3. The process of detecting DNA by using the SERS liquid-phase chip method is the same as that described in Examples 1-3.
[0061] A detection limit of 10 was obtained -11 M.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com