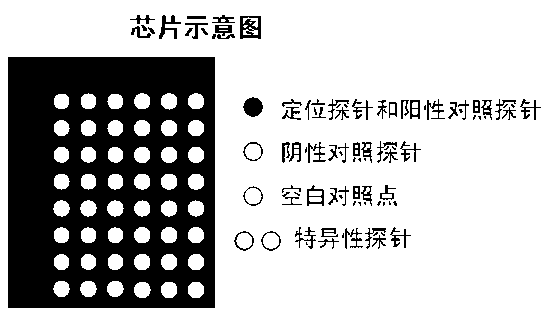
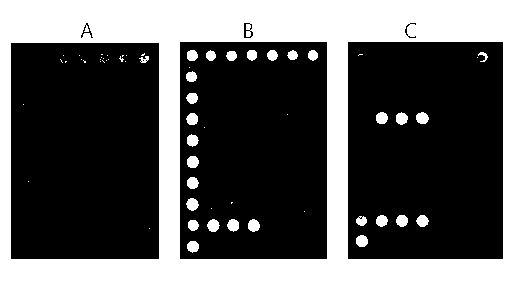
Gene chip-based method for synchronously and rapidly detecting thirteen pathogenic microorganisms in water body
A technology for pathogenic microorganisms and detection methods, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, and resistance to vector-borne diseases. It can solve the problems of long time, amplification failure, non-specific products, etc. High specificity, simple operation and strong stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0050] Taking the drinking water source water sources in the Three Gorges Reservoir area of the Yangtze River as the detection object, the water outlets of the five tap water plants in the Chongqing section of the Yangtze River Basin are selected for the water samples for water samples.The discharge port and landscape waters are collected. This method is used for detection. The water sample information is shown in the table below.
[0051] Table 3 water sample information
[0052] serial number Water sample name Collection point Collection location description 1 Yangtze River Drinking Water Source Water 1 A water plant takes water points At 500m upstream, the water flow by the river is smooth, and the water is taken at 20cm underwater 2 Changjiang Drinking Water Source Water 2 Take a water point for a drink station At 500m upstream, the water flow by the river is smooth, and the water is taken at 20cm underwater 3 Changjiang Drinking Water Source Wat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com