Construction and application of cell model for cotransfection of drug metabolic enzyme and transporter
A cell model and metabolic enzyme technology, which is applied in the field of co-transfection of drug-metabolizing enzyme CYP3A4 and transporter MDR1 cell model, can solve the problem of no stable co-transfection cell model, etc., and achieve the effect of short culture period and easy culture
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
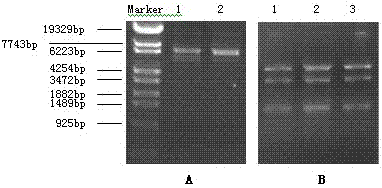
[0034] Identification of expression plasmid pcDNA3.1(+) / MDR1 and construction of pcDNA3.1(+) / Hygro / CYP3A4 recombinant plasmid
[0035] The plasmid pcDNA3.1(+) / MDR1 was used as the expression vector, and the competent bacteria E.coli DH 5α was used to amplify the plasmid, and the QIAprep Spin Miniprep kit was used to extract and purify the plasmid. The full length of MDR1 in / MDR1 was sequenced by DNA. Design PCR primers containing Kpn I and Xho I restriction sites, the upstream and downstream primer sequences are 5'-CGGGGTACCATGGCTCTCATCCCAGACTTGGC-3' respectively;
[0036] 5'-ATCTCGAGTCAATGATGATGATGATGATGGGCTCCACTTACGGTGC-3'.
[0037] The CYP3A4 gene was fished from the gene bank of our laboratory, and T-A was connected to pMD19-T Vector. After transformation, bacterial picking, shaking, and plasmid extraction, enzyme digestion and sequencing were performed. Kpn I and Xho I double-digested pMD9-T / CYP3A4 to obtain CYP3A4 cDNA with cohesive ends, which was ligated with the pc...
Embodiment 2
[0039] Example 2 MDCK cell transfection and functional identification
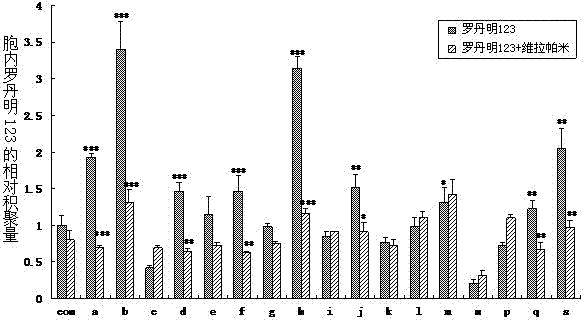
[0040] MDCK cells were treated with 10 5 Seed in a six-well plate per well and culture for 48 hours. With Lipofectamine TM The 2000 reagent transfects the expression vector pcDNA3.1(+) / MDR1 into MDCK cells. For the method, refer to the instruction manual of the transfection reagent. After 24 hours of transfection, the medium was changed, and G418 was added for selection. The concentration was 600 μg / ml for 96 hours and then increased to 800 μg / ml for 24 hours. The surviving cells were then transferred to culture flasks for culture at a concentration of 600 μg / ml for about 20 days. Inoculated in 96-well plates according to the limiting dilution method, and obtained 19 resistant monoclonals, which were MDCK-MDR1 cells, named after lowercase letters a~s.
[0041] 1. The accumulation of Rho123 in cells
[0042] MDCK and MDCK-MDR1 cells in 2×10 5 Seed each well in a 24-well plate. After 48 hours, add DME...
Embodiment 3
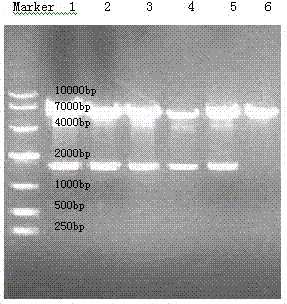
[0073] Example 3 Transfection of MDCK-MDR1 cells with pcDNA3.1(+) / Hygro / CYP3A4
[0074] MDCK-MDR1 cells were seeded in 6-well plates at the same density and transfected when the cells reached 50-70% confluency. Using Lipofectamine TM The LTX transfection reagent transfects the expression vector pcDNA3.1(+) / Hygro / CYP3A4 into MDCK-MDR1 cells. For the method, refer to the instructions of the transfection reagent. After 6 hours of transfection, the DMEM medium containing 10% FBS was replaced. After 24 hours of cultivation, the medium was replaced with DMEM medium containing 400 mg / L hygromycin B and 10% FBS, and the medium was changed every other day. The surviving cells were inoculated in 96-well plates according to the limiting dilution method about 14 days after drug addition and screening, and resistant monoclonals were obtained, which were respectively named P3.1 and P4 series with Arabic numerals.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com