Method for the preparation of diols
A technology of aliphatic diols and microorganisms, which is applied in the directions of botany equipment and methods, biochemical equipment and methods, applications, etc., and can solve problems such as expensive production processes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach
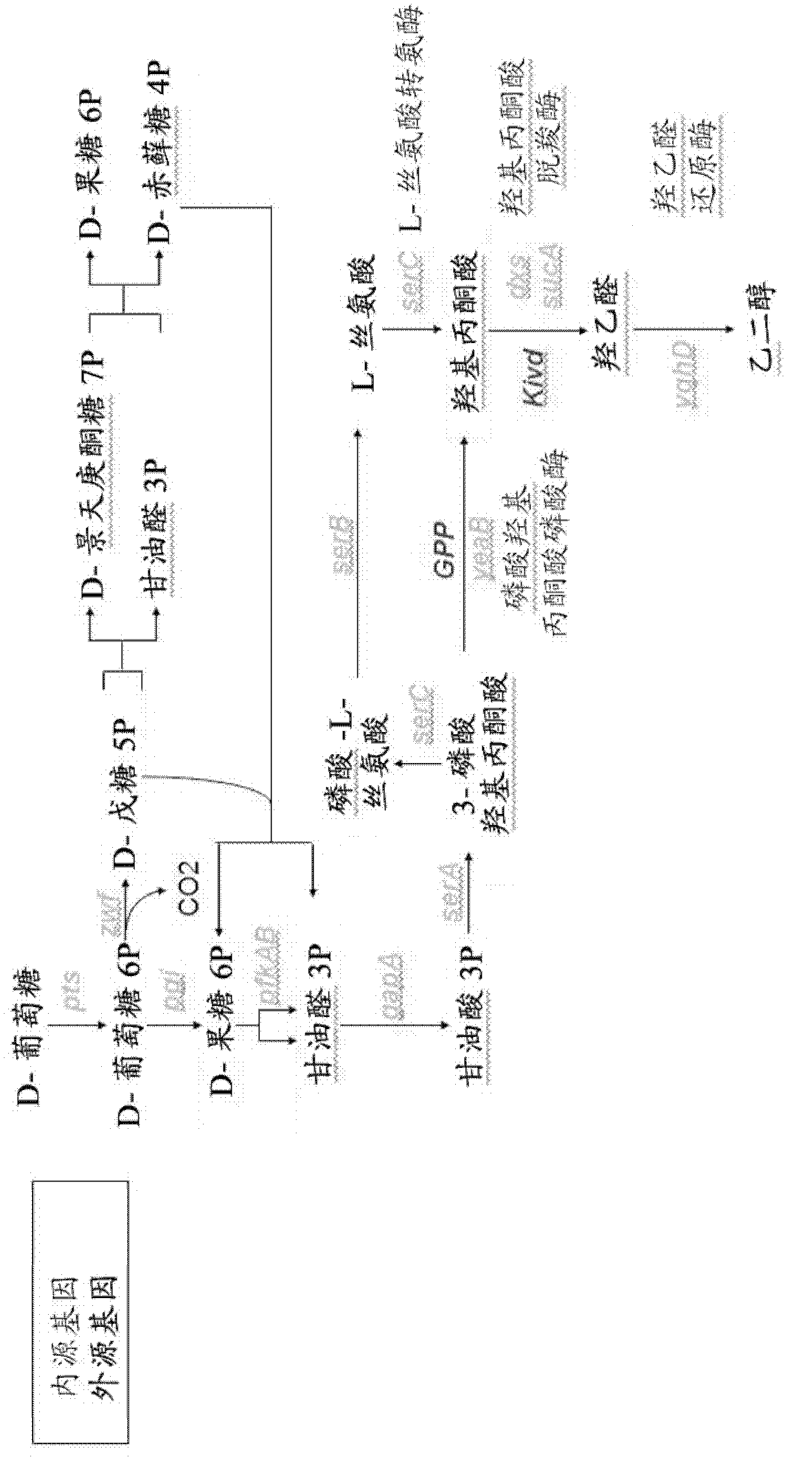
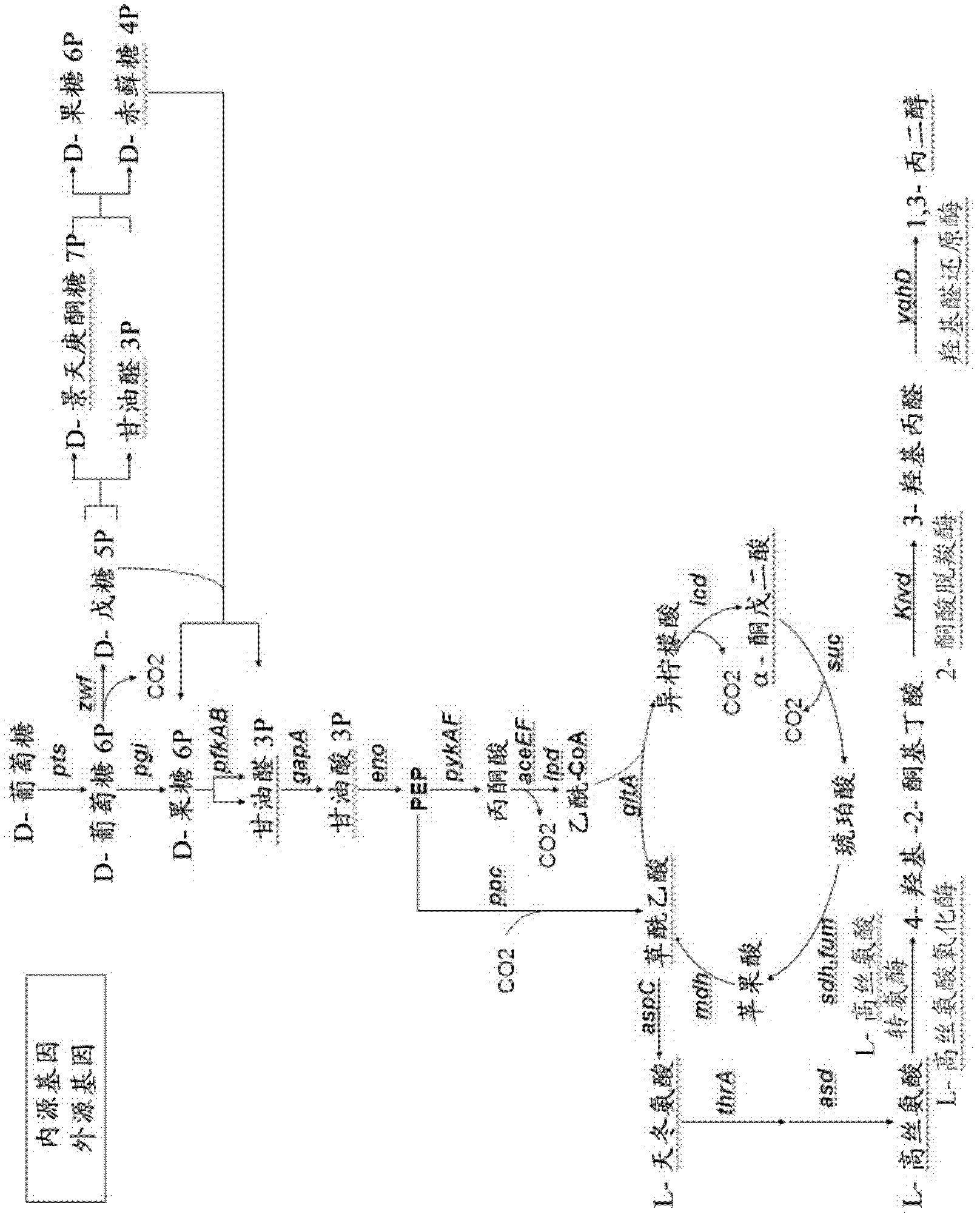
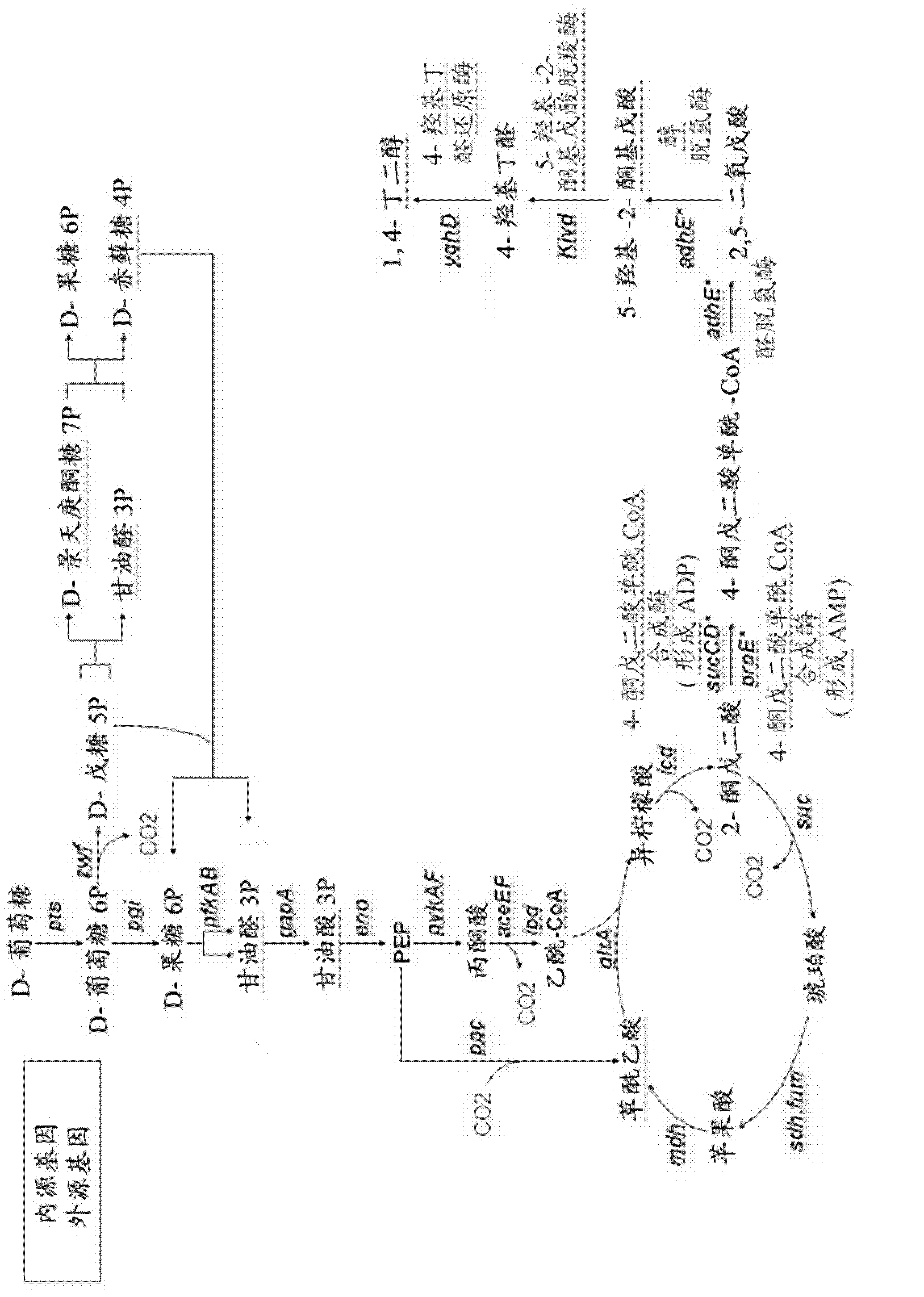
[0064] Other embodiments of the present invention will be described below. The microorganisms are modified to favor the production of hydroxy-2-keto-aliphatic acid metabolites and to convert the products obtained from the decarboxylation step of the same hydroxy-2-keto-aliphatic acid metabolites to the corresponding aliphatic diols.
[0065] The following description is for E. coli, a microorganism lacking endogenous 2-ketoacid decarboxylase activity. Thus, a heterologous gene encoding this activity is introduced into the microorganism.
[0066]The microorganism is modified to optimize the pathway for producing a hydroxy-2-ketoaliphatic acid metabolite and to convert the product obtained from the decarboxylation step of the same hydroxy-2-ketoaliphatic acid metabolite to an aliphatic diol or Based on the known metabolic pathways and endogenous genes of E. coli. However, those skilled in the art can use similar strategies to introduce or delete corresponding genes in other mi...
Embodiment 1
[0106] Construction of a strain expressing the gene encoding 2-ketoacid decarboxylase and the gene encoding hydroxyaldehyde reductase: MG1655 (pME101-kivDll-yqhD-TT07)
[0107] 1.1 Construction of plasmid pM-Ptrc01-kivD11-TT07 for overexpressing kivD of Lactococcus lactis encoding α-ketoisovalerate decarboxylase:
[0108]The synthetic gene of Lactococcus lactis kivD encoding α-ketoisovalerate decarboxylase was prepared by Geneart (Germany). The codon usage and GC content of this gene were adapted to E. coli according to the supplier matrix. Expression of this synthetic gene is driven by the constitutive Ptrc promoter. A transcription terminator was added downstream of the gene. Constructs were cloned into the supplier's pM vector and verified by sequencing. If necessary, the synthetic gene was cloned into pME101 vector (this vector was derived from plasmid pCL1920 (Lerner & Inouye, 1990, NAR 18, 15p 4631)), which was then used to transform E. coli strains.
[0109] Ptrc0...
Embodiment 2
[0152] Construction of strains with increased flux in the ethylene glycol pathway: MG1655ΔsdaAΔsdaB ΔpykFPtrc 18-gpmA Ptrc 18-gpmB(pME 101-kivDll-yqhD-TT07)
[0153] 2.1 Construction of MG1655ΔsdaAΔsdaB strain
[0154] To delete the sdaA gene, the homologous recombination strategy described by Datsenko & Wanner (2000) was used. This strategy allows the insertion of chloramphenicol or kanamycin resistance cassettes while deleting most of the genes involved. For this purpose the following oligonucleotides were used:
[0155] ΔsdaAF (SEQ ID NO 10)
[0156] gtcaggagtattatcgtgattagtctattcgacatgtttaaggtggggattggtccctcatcttcccataccgtagggccTGTAGGCTGGAGCTGCTTCGhas
[0157] - and the sequence of the sdaA gene (1894941-1895020) (see the website for the reference sequence http: / / genolist.pasteur.fr / Colibri / ) homologous region (lowercase),
[0158] - a region for amplification of the kanamycin resistance cassette (see Datsenko, K.A. & Wanner, B.L., 2000, PNAS, 97: 6640-6645 for refere...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com