A method for constructing t vector
A carrier and precursor technology, applied in the field of genetic engineering, can solve the problems of many quality control links, low quality of the T carrier, and cumbersome production process, reducing operation steps and control links, high enzyme digestion yield, and self-cyclization. low rate effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Embodiment 1: Amplification of the resistance gene fragment containing AhdI site
[0042] Using the pEGFP-C1 plasmid as a template, use the upstream primer with the nucleotide sequence shown in SEQ ID No: 2 and the downstream primer with the nucleotide sequence shown in SEQ ID No: 3 to carry out PCR amplification to obtain the The kanamycin resistance gene fragment at the AhdI site.
[0043] The PCR amplification reaction system is:
[0044]
[0045] Add sterilized distilled water to make up the total amount of the system to 20 μL.
[0046] The PCR reaction procedure is:
[0047]
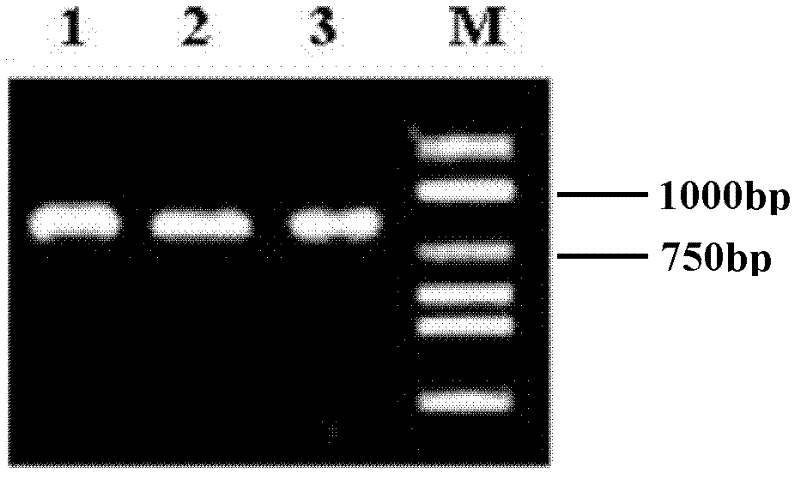
[0048] The resulting PCR product was purified and recovered, and 5 μL of 1% agarose gel electrophoresis was used for detection. The results are shown in figure 1
Embodiment 2
[0049] Example 2: Preparation of T carrier precursor
[0050] The PCR product recovered in Example 1 and the pUC-19 plasmid were double-digested with BamHI and HindIII respectively (the endonuclease and corresponding buffer used were Takara products), and the reaction system was as follows:
[0051] Add the following ingredients to Tube A:
[0052]
[0053] Add sterilized distilled water to make up the total amount of the system to 30 μL.
[0054] Add the following ingredients to tube B:
[0055]
[0056] Add sterilized distilled water to make up the total amount of the system to 30 μL.
[0057] Put tubes A and B in a constant temperature water bath at 37°C for 6-8 hours for digestion and digestion, and then perform gel electrophoresis and DNA gel recovery for the digested products. The recovered fragments were ligated overnight at 16°C under the action of T4 DNA ligase (8-12h).
[0058] Connection reaction system:
[0059]
[0060]
[0061] Add sterilized dis...
Embodiment 3
[0063] Embodiment 3: Preparation of T carrier
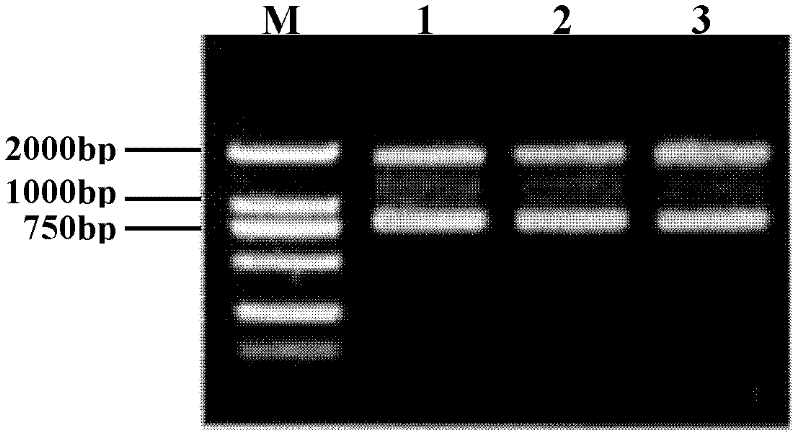
[0064] The T vector precursor obtained in Example 2 was digested with the specific restriction endonuclease AhdI in the following manner, and the linear vector with a size of about 2000 bp was recovered by electrophoresis, which was the T vector.
[0065] Enzyme digestion reaction system:
[0066]
[0067] Add sterilized distilled water to make up the total amount of the system to 30 μL.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com