Molecule marking method for identifying tilapia nilotica, oreochromis aureus and hybridized fish thereof
A Nile tilapia and molecular marker technology, applied in biochemical equipment and methods, microbiological determination/inspection, etc., can solve the problems of amplification band change, sensitive reaction conditions, poor stability and repeatability, etc., and achieve The effect of reducing detection time, reducing detection cost and stabilizing reaction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
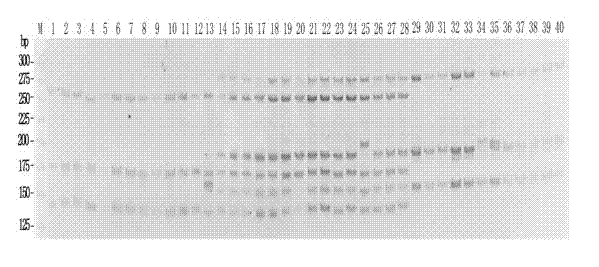
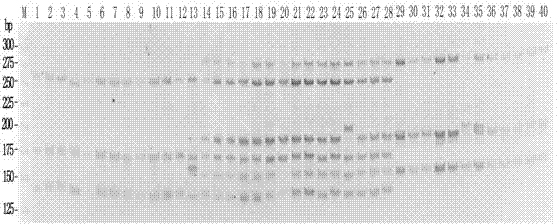
[0038] A kind of molecular marker method for distinguishing Nile tilapia, Olia tilapia and hybrid fish thereof of the present invention adopts the following technical steps:
[0039] Thirteen Nile tilapia were collected from Yixing Fishery, Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences. 0.5 mL of blood was collected from the tail vein of each fish, and 30 μL of blood cells were taken to extract genomic DNA. Using the genomic DNA of Nile tilapia as a template, three primer pairs of primers GM017, UNH948 and GM440 were used for multiplex PCR amplification. The total volume of the PCR reaction was 15 μL, which contained 1.5 μL of 10× reaction buffer, MgC1 2 2 mmol / L, dNTP 200 μmol / L, 3 pairs of upstream and downstream primers each 0.2 μmol / L, Taq Enzyme 0.4U, DNA 80 ng, make up volume with sterilized double distilled water. The PCR reaction conditions were: 94°C for 3 min; 94°C for 30 s, 52°C for 30 s, 72°C for 30 s, 25 cycles; 72°C for 8 min. Aft...
Embodiment 2
[0045] A kind of molecular marker method for distinguishing Nile tilapia, Olia tilapia and hybrid fish thereof of the present invention adopts the following technical steps:
[0046] 8 Aoni hybrid tilapia (Nile tilapia ♀ × Olia tilapia ♂) were collected from Yixing Fishery, Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences. 0.1 g of caudal fin rays were taken from each fish, and genomic DNA was extracted. Using the genomic DNA of Oni hybrid tilapia as a template, multiplex PCR amplification was performed with 3 primer pairs of primers GM017, UNH948 and GM440. The total volume of the PCR reaction was 15 μL, which contained 1.5 μL of 10× reaction buffer, MgC1 2 2 mmol / L, dNTP 200 μmol / L, 3 pairs of upstream and downstream primers each 0.2 μmol / L, Taq Enzyme 0.4U, DNA 50 ng, make up volume with sterilized double distilled water. The PCR reaction conditions were: 94°C for 3 min; 30 cycles of 94°C for 30 s, annealing at 52°C for 30 s, and 72°C for 30 s; ...
Embodiment 3
[0052] A kind of molecular marker method for distinguishing Nile tilapia, Olia tilapia and hybrid fish thereof of the present invention adopts the following technical steps:
[0053] 12 Olia tilapia and 7 Neo hybrid tilapia (Nile tilapia♂×Olia tilapia♀) were collected from Yixing Fishery, Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences. Genomic DNA was extracted from 30 μL of blood cells from each fish. Using these genomic DNAs as templates, multiplex PCR amplification was performed with 3 primer pairs of primers GM017, UNH948 and GM440. The total volume of the PCR reaction was 15 μL, which contained 1.5 μL of 10× reaction buffer, MgCl 2 2 mmol / L, dNTP 200 μmol / L, 3 pairs of upstream and downstream primers each 0.2 μmol / L, Taq Enzyme 0.4U, DNA 70 ng, make up volume with sterilized double distilled water. The PCR reaction conditions were: 94°C for 3 min; 94°C for 30 s, 52°C for 30 s, 72°C for 30 s, 28 cycles; 72°C for 8 min. After the PCR reaction...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com