Method for detecting and counting live microbes
A technology of microorganisms and target microorganisms, which is applied in the field of inspection and counting of living microorganisms, can solve the problems of poor specificity of inspection and counting methods, time-consuming and laborious identification, etc., and achieve the effect of strengthening the detection of probiotics, short operation time and good reliability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Bifidobacterium longum (Bifidobacterium longum) count in embodiment 1 bifidobacterium yoghurt
[0034] 1. Design of Bifidobacterium longum qualitative PCR reagent
[0035] Sequence the gene sequence of the region between 16S rRNA and 23S rRNA of the fermented Bifidobacterium longum strain, compare and analyze on NCBI online, design Bifidobacterium longum qualitative PCR primers, upstream primers: 5'-CCA TCA TCC GCT TTC G-3', see SEQ ID NO.1 in the sequence listing; downstream primer: 5'-TGG CAG ACA GGA CCG ATG-3', see SEQ ID NO.2 in the sequence listing. The PCR product length is expected to be 215bp.
[0036] The bacterial positive marker PCR primers are 16S rRNA universal primers, upstream primers: 5'-AGA GTTTGA TCC TGG CTC AG-3'; downstream primers: 5'-C TGC TGC CTC CCG TAGGAG-3'. The length of the PCR product is expected to be about 300bp.
[0037] 2. Plate counting, PCR identification of colonies.
[0038] Aseptically weigh 25g of bifidobacterium yogurt (Changy...
Embodiment 2
[0046] Bifidobacterium longum live bacteria count in the intestinal tract of embodiment 2
[0047] 1. Bifidobacterium longum qualitative PCR reagent
[0048] With embodiment 1.
[0049] 2. Plate counting, PCR identification of colonies
[0050] Aseptically weigh fresh human feces, blow in carbon dioxide gas, oscillate and mix with sterile water with glass beads, dilute, and count on plates with TPY agar medium, the counting result is 6.3×10 9 cfu / g. Other steps are with embodiment 1.
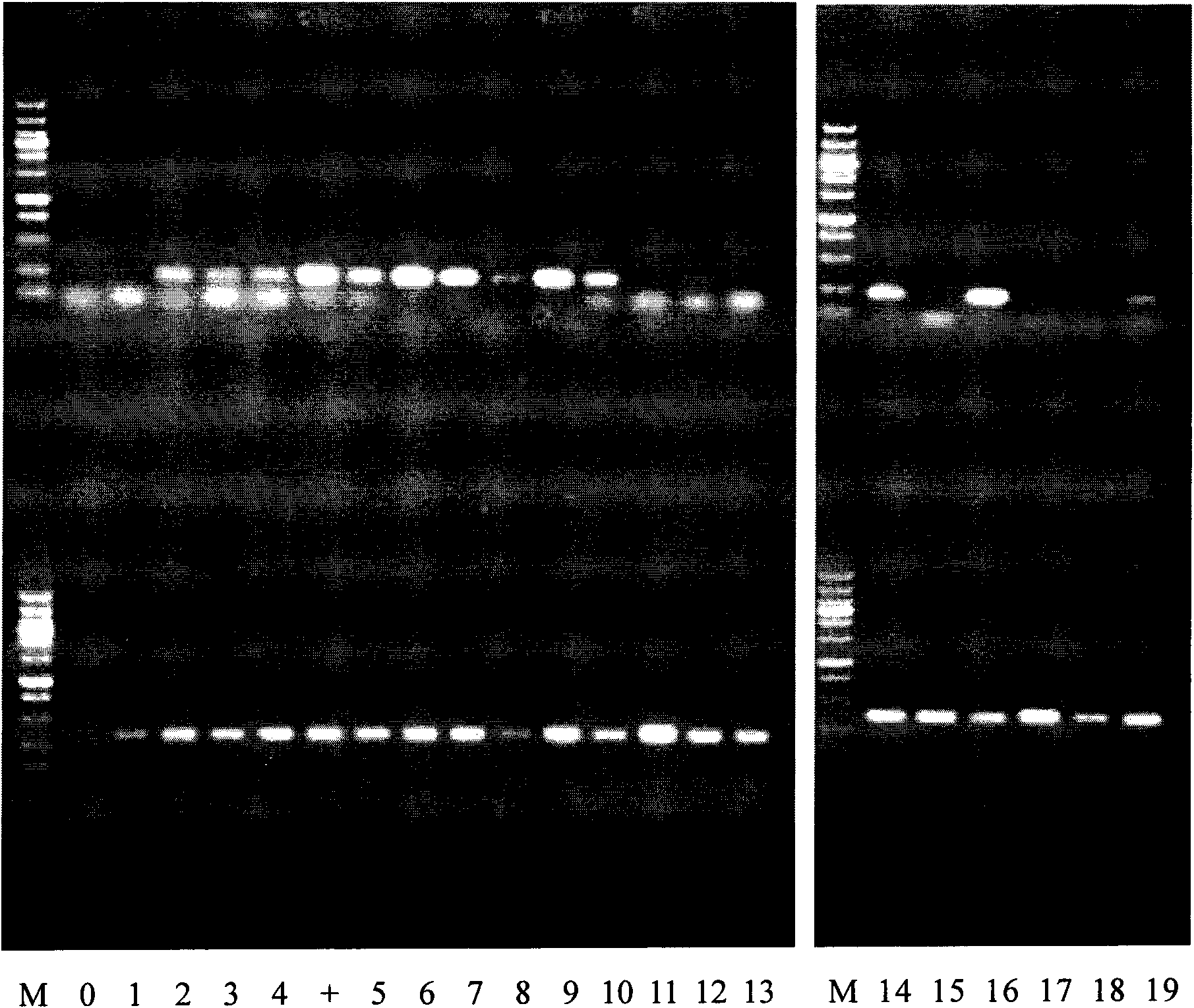
[0051] The agarose electrophoresis diagram of the PCR product is shown in figure 2 , the upper band in the figure is the qualitative PCR band of Bifidobacterium longum with the corresponding number, and the lower part is the PCR band of the positive marker of the bacteria with the corresponding number, the number M is Wide Range DNA Marker, the number 0 is the negative control, and the number + is positive control, figure 2 Among them, 1, 2, 3...19 represent the selected single colonies ...
Embodiment 3
[0054] Detection and counting of common heat-resistant bacillus in embodiment 3 milk
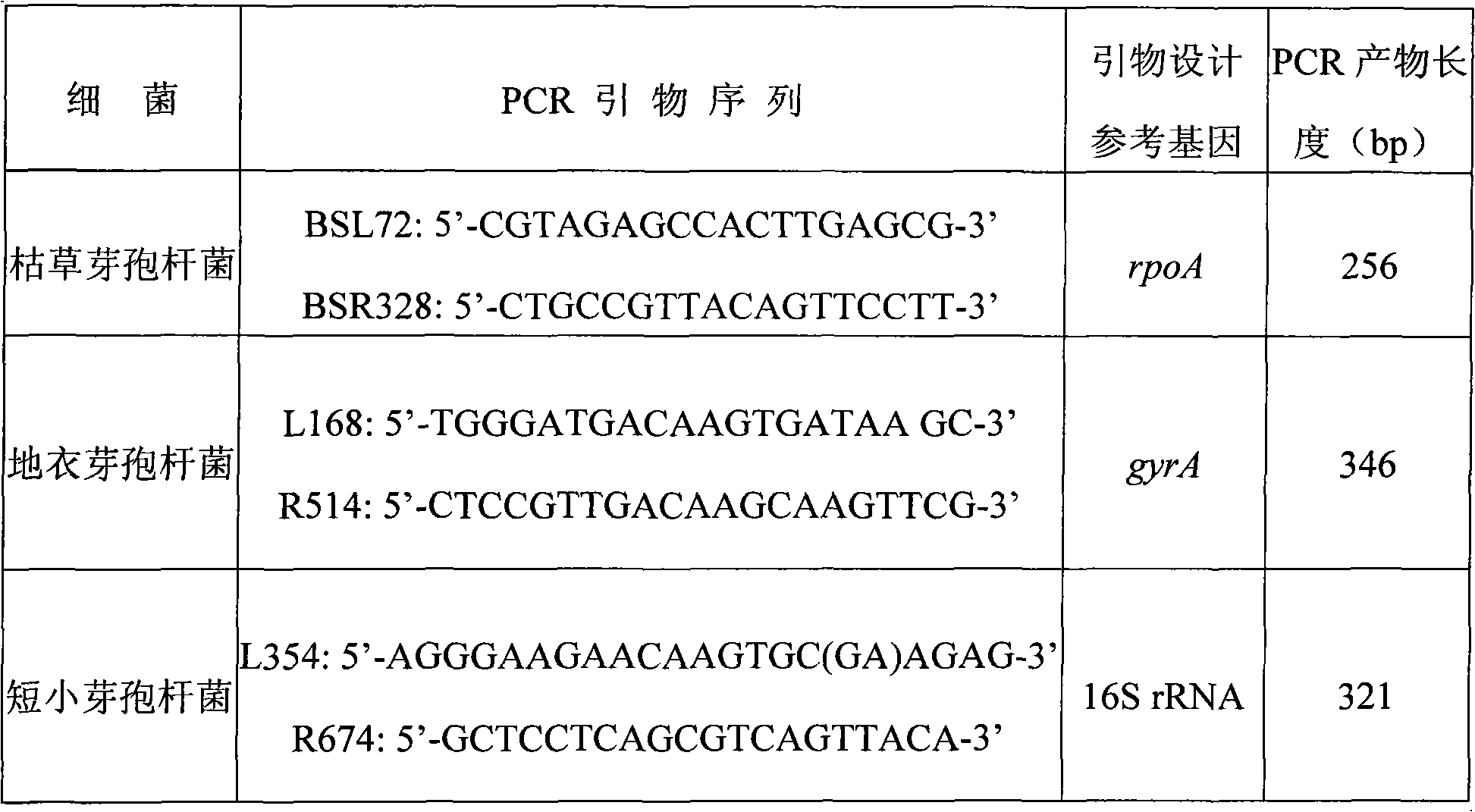
[0055] Bacillus subtilis, B. licheniformis and B. pumilus in group Bacillus subtilis are common heat-resistant bacillus in milk, and they affect the quality of milk products. The specific multiplex PCR primer sequence comes from the literature "Application of Multiplex PCR to Identify Bacillus Commonly Used in Microbial Fertilizers" (Cao Fengming, Shen Delong, Li Jun, etc. Acta Microbiology, 2008, 48(5): 651-656), see Table 1; Bacteria positive Marker PCR primers are the same as in Example 1.
[0056] Table 1. PCR Primers
[0057]
[0058] Fresh milk extruded from cows that has just been purchased is treated at 85°C for 10 minutes to kill non-heat-resistant bacteria and enrich heat-resistant bacillus. Use pre-sterilized saline test tubes to make 10-fold serial dilutions, and the selected dilution factor is 10 1 、102 、10 3 Take 1ml of the test tube and put it into a sterile petri dish,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com