Molecular marker identification method of black fungus strain and specific molecular marker primers of black fungus strain HW 5
A technology of molecular markers and identification methods, which is applied in the fields of biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc. Large and other problems, to achieve the effect of ensuring specificity, strong specificity, and small screening amount
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
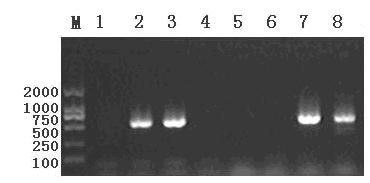
[0012] Specific embodiment one: the molecular marker identification method of black fungus strains in this embodiment is realized by the following steps: one, extract the total DNA of selected black fungus strain mycelium; two, ISSR-PCR analysis: with selected black fungus strain mycelium The total DNA is used as a template, and the ISSR primers are screened to obtain specific molecular markers; 3. Purify the specific molecular markers, and design specific molecular marker primers; The black fungus bacterial strain is selected black fungus bacterial strain, and the black fungus bacterial strain that does not obtain positive band is other black fungus bacterial strains; Wherein the specific molecular mark that step 2 obtains exists in a nucleus of selected black fungus bacterial strain mycelium, and not Exist in another nucleus of the mycelia of selected black fungus strains.
specific Embodiment approach 2
[0013] Embodiment 2: The difference between this embodiment and Embodiment 1 is that in step 1, the plant genome DNA extraction method is used to extract the total DNA of the selected black fungus strain mycelium. Other steps and parameters are the same as those in Embodiment 1.
specific Embodiment approach 3
[0014] Specific embodiment three: the molecular marker identification method of black fungus strains in this embodiment is realized according to the following steps: one, extract the total DNA of black fungus strain HW5 mycelia; two, ISSR-PCR analysis: use the total DNA of black fungus strain HW5 mycelia As a template, carry out ISSR primer screening to obtain specific molecular markers; 3. Purify specific molecular markers and design specific molecular marker primers; 4. Use specific molecular marker primers to perform PCR detection on black fungus strains to be tested to obtain black fungus with positive bands The bacterial strain is the selected black fungus strain, and the black fungus strain that does not get the positive band is other black fungus strains; wherein the specific molecular marker obtained in step 2 exists in a nucleus of the selected black fungus strain mycelium, and does not exist in In another nucleus of the mycelia of selected black fungus strains.
[00...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com