Method for fast detection and identification of bacterial wilt caused by infection of ralstonia solanacearum
A technology for identification of solanaceae solanaceous wilt bacteria, which is applied in the field of rapid detection and identification of solanaceous wilt, can solve the problems of rare plant pathogenic microorganisms, no identification of solanaceae solanaceae, and achieves accurate, reliable and adaptable identification results. Strong, low-cost effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1: Detection and identification of known disease samples
[0033] 1. Experimental materials: Eggplant and tomato disease-like tissues caused by the infection of R. solanacearum and known healthy eggplant tissues were used as materials.
[0034] 2. Experimental method:
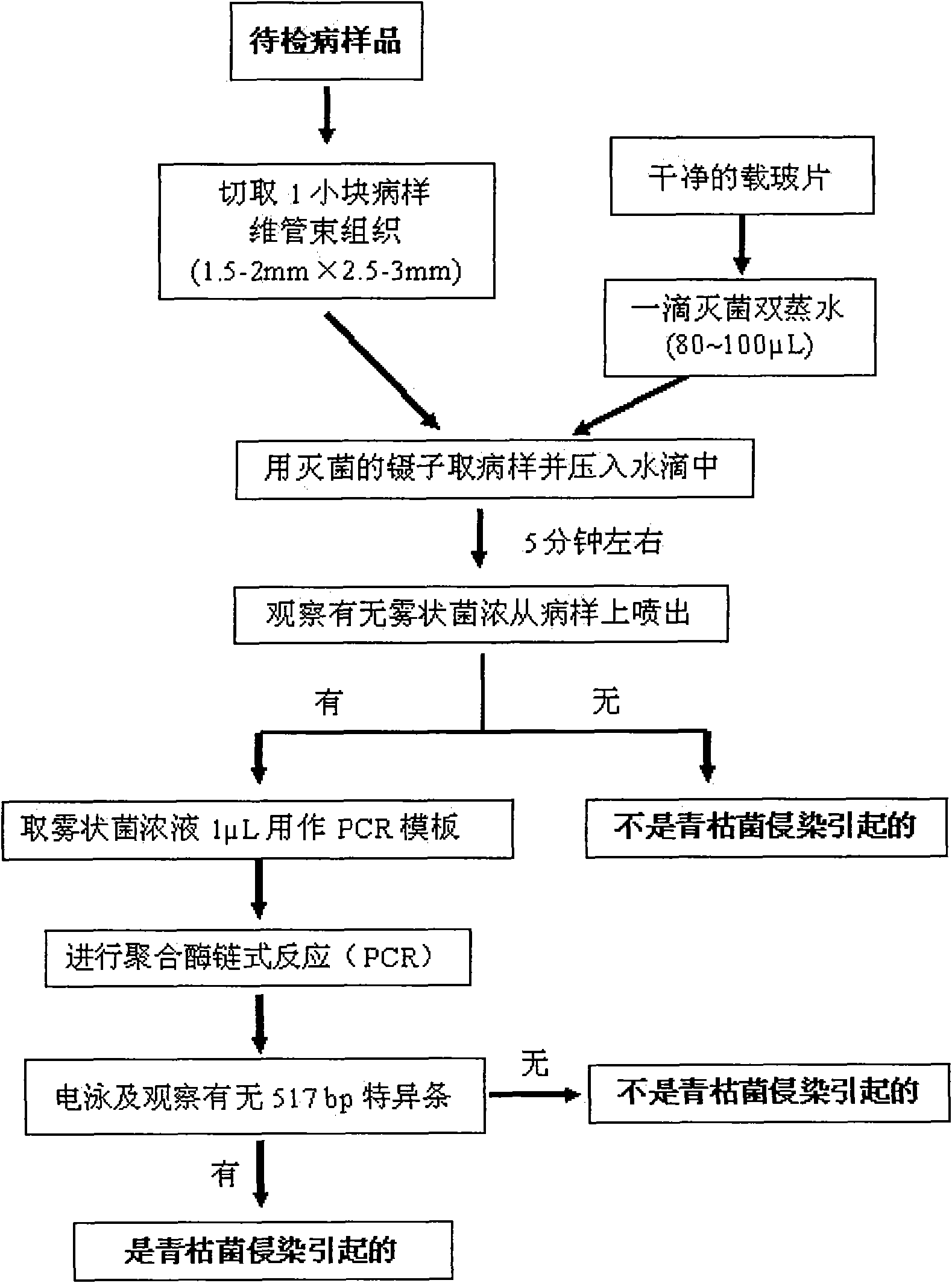
[0035] (1) Rinse the stem base and root of the diseased sample to be tested with water, and take a vascular bundle tissue with a size of about 1.5-2 mm × 2.5-3 mm from the stem base of the diseased sample to be tested, and 8 samples to be tested include 4 eggplants diseased sample and 4 tomato diseased samples, and another healthy eggplant sample was taken as a control;
[0036] (2) Use a sterile pipette to draw 100 μL of sterilized water, drop it on a clean glass slide, and use sterilized tweezers to put the diseased tissue into the 100 μL sterilized water drop on the clean glass slide. Forcefully press the diseased tissue into the water droplets, and let it stand for about 5 minutes; observe ...
Embodiment 2
[0042] Embodiment 2: detection and identification test to unknown disease sample
[0043] 1. Experimental materials: 10 different plant bacterial wilt samples or asymptomatic healthy samples collected from fields in multiple regions were tested.
[0044] 2. Experimental method:
[0045] 10 different plant samples collected from multiple regions in Guangdong were sampled, and a total of 17 samples were obtained and numbered. The plant species, collection locations, and symptoms corresponding to each number of samples are shown in Table 1. The method used for detection and identification is the same as that used in Example 1, and is compared and verified with the traditional method for isolation and identification of plant pathogenic bacteria.
[0046] The specific steps of the traditional method for isolation and identification of plant pathogenic bacteria include: 1. Disinfect the surface of the diseased tissue to be examined with 75% alcohol; Streak and culture at 28-30°C f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com