Method and material for simultaneously detecting mycobacterium tuberculosis compound and identifying its medicament resistance
A technology of mycobacterium tuberculosis and mycobacteria, applied in the fields of molecular biology, medicine, and microbiology, can solve the problems of expensive, difficult to prepare probe sets, and inability to detect the sensitivity of pathogens, achieving short-term and low-cost Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
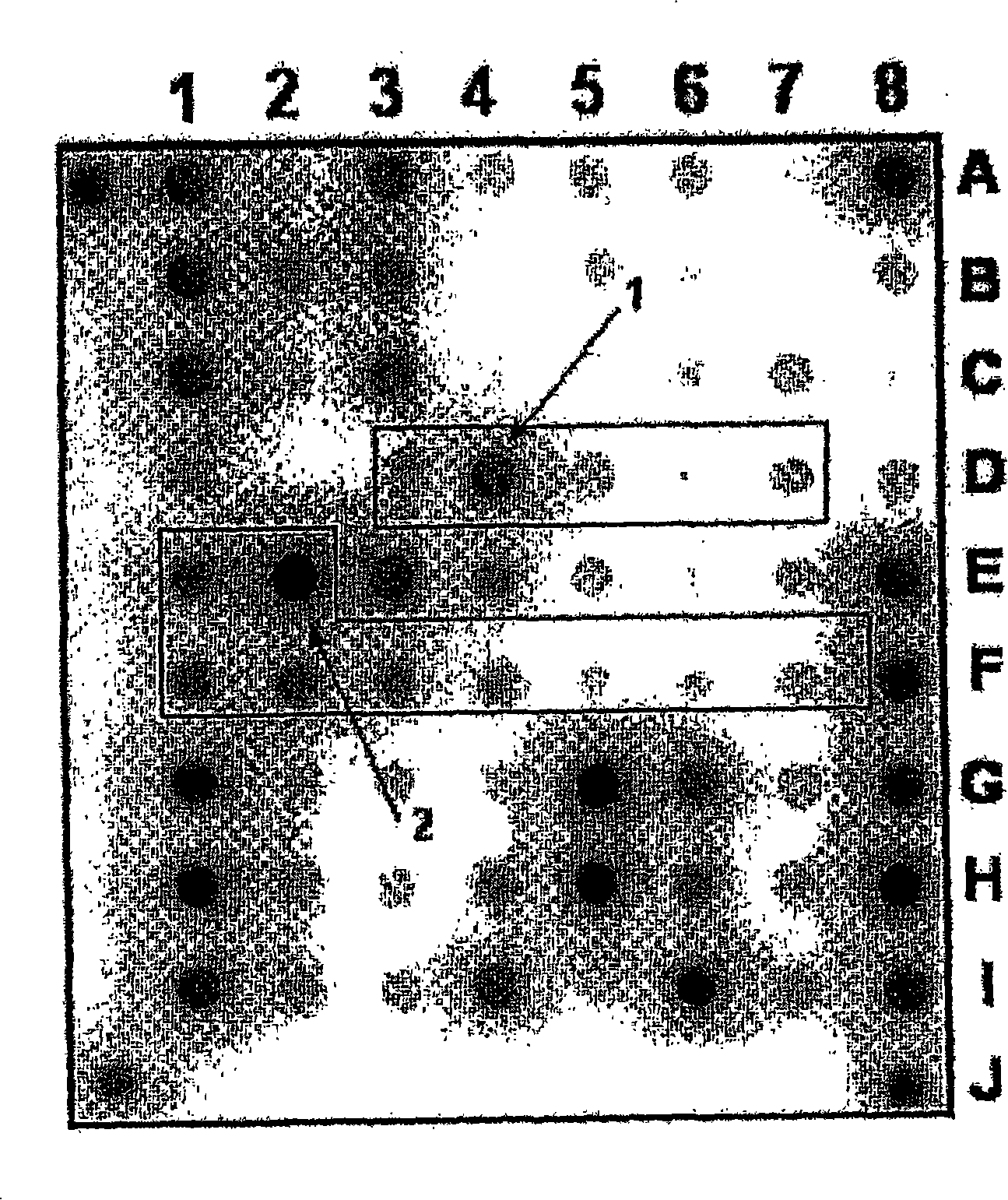
[0123] Example 1. Biochip for detection of Mycobacterium tuberculosis complex and evaluation of strain sensitivity to rifampicin and isoniazid
[0124]1. The oligonucleotides for immobilization on the biochip and the primers for amplification are synthesized on the automatic synthesizer 394DNA / RNA synthesizer (Applied Biosystems, USA), and contain the oligonucleotides for further immobilization on the gel respectively. The free amino 3′-amino-modifier C7 CPG 500 (Glen Research, USA) on the fluorochrome or the spacer for the 5′-amino-modifier C6 (Glen Research, USA) for attaching the fluorescent dye. Attachment of indodicarbocyanine fluorescent dye ("Biochip-IMB", Russia) was performed according to the manufacturer's recommendations. Biochips were produced according to methods described earlier (Rubina AY, Pan'kov SV, Dementieva EI et al., Hydrogel drop microchips with immobilized DNA: properties and methods for large-scale production. AnalBiochem 2004; 325: 92-106). The bioch...
Embodiment 2
[0143] Example 2. Processing of clinical samples
[0144] 1. Mix clinical samples (sputum, exudates, eluate, bronchoalveolar lavage) with a freshly prepared 0.5% solution of N-acetyl-L-cysteine (NALC) in 2% NaOH Mix in a 1:1 (v / v) ratio. The samples were stirred vigorously with a Vortex and maintained at room temperature for 20 minutes. Phosphate buffered saline pH 6.8 was added to the sample at a ratio of 1:5 (v / v), and the mixture was centrifuged at 3,000 rpm for 30 minutes. When using cerebrospinal fluid, perform a preliminary centrifugation at 10,000 rpm for 10 minutes. For blood analysis, the lymphocyte fraction was initially isolated according to the usual method using Ficoll. Subsequent processing was the same for all samples.
[0145] 2. The precipitated pad was suspended with 1.5 mL of TE buffer (10 mM Tris-HCl, 1 mM EDTA) (pH 8.0), and then centrifuged at 3,000 rpm for 30 minutes. Repeat the washing process several times.
[0146] 3. Add 30 μl of TE buffer (p...
Embodiment 3
[0148] Example 3. Amplification of fragments of IS6110 movable factor, rpoB, katG, inhA, ahpC genes; preparation of single-stranded fluorescently labeled fragments by multiplex PCR
[0149] In the first step, multiplex amplification of fragments of the rpoB (212 b.p.), katG (166 b.p.), inhA (133 b.p.), ahpC (126 b.p.) genes and the IS6110 mobile factor (309 b.p.) was performed.
[0150] 3 μl of the sample obtained in Example 2, paragraph 4, were added to 25 μl of the PCR-mix.
[0151] Composition of the PCR-Mix:
[0152] 1X PCR-buffer: 10 mM KCl, 10 mM Tris-HCl (pH 8.3) (Sileks, Russia);
[0153] 1.5mM MgCl 2 ;
[0154] dATP, dCTP, dGTP, and dUTP (Sileks) are each 200 μM;
[0155] A mixture of primers (sequences listed in Table 2) at the following concentrations: p105f, p293r, katG_f, katG_r1, IS_f, IS_r1; InhA_f, InhA_r1, ahpc_f, ahpC_r1 - 100 nM;
[0156] 5U thermostable Taq DNA polymerase (sileks);
[0157] • 0.5 U uracil-DNA-glycosylase (Sileks).
[0158] Amplify on...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com