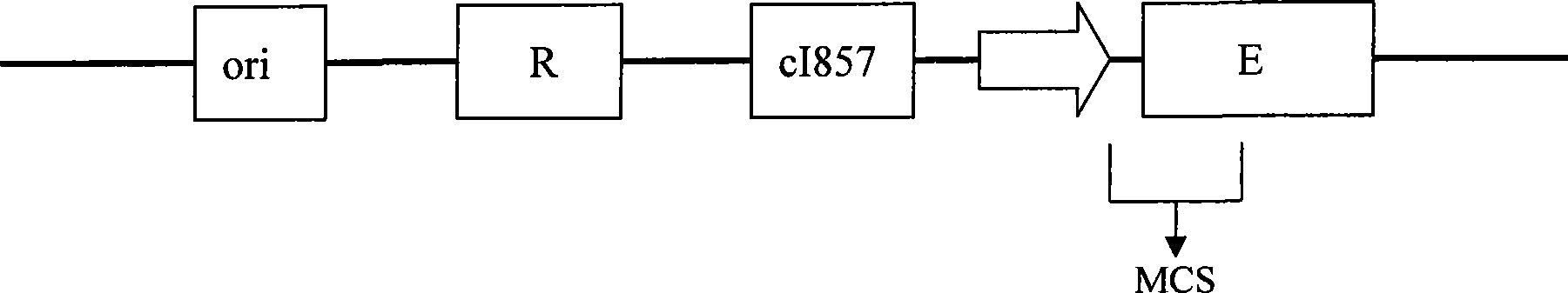
Design and construction method of high efficiency temperature controlled positive screening cloning carrier
A cloning vector and positive screening technology, which can be used in the introduction of foreign genetic material using vectors, recombinant DNA technology, etc., and can solve the problems of cumbersome operation, time-consuming and laborious
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Implementation Example 1: Construction of common cloning vector pClonEZ
[0025] Taking the non-patented plasmid vector pBV220 widely used in domestic laboratories as the starting vector, it was constructed according to the following steps and methods:
[0026] 1. Using the genomic DNA of the commercially available phage Φ×174 as a template, the complete E gene coding sequence (273 bp) was amplified by PCR technique. When designing primers for amplifying the E gene, an EcoRI site was introduced into the upstream primer, a PstI site was introduced into the downstream primer, and 4 protective bases were introduced outside the restriction site. PCR amplification was carried out according to the selected high-fidelity Taq enzyme product manual and the Tm value of the primers. The size of the amplified product is about 300bp.
[0027] 2. Use EcoRI and PstI to double-digest plasmid pBV220 and the above PCR-amplified E gene fragment respectively, and tap rubber to recover th...
example 2
[0036] Example 2: Construction of T vector pEZ-T
[0037] The T vector pEZ-T for T-A cloning is constructed with the pClonEZ constructed in Example 1 as a starting vector:
[0038] 1. Use the commercially available QuikChange(R) site-directed mutagenesis kit and carry out point mutations to pClonEZ according to the method described in its product manual, and mutate the EcoR I site sequence before the E gene into the EcoR V site sequence, and the resulting plasmid is pEZ-T pre-carrier.
[0039] 2. Digest the above pEZ-T pro-vector with EcoR V, and the enzyme digestion reaction system and conditions are carried out according to the corresponding endonuclease product instructions.
[0040] 3. Purify and recover the linearized pEZ-T procarrier by conventional phenol-chloroform extraction and ethanol precipitation methods.
[0041] 4. Using the above-mentioned pEZ-T pro-vector and dTTP as substrates, use Taq DNA polymerase to carry out terminal T addition reaction, and the reacti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com