Isolation and expression of a gene for a nitrilase from acidovorax facilis 72W
A nitrilase and chimeric gene technology, applied in the field of molecular biology, can solve the problems of lack of thermostable high-yield nitrilase, thermal instability limiting industrial application and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0196] Purified nitrilase protein
[0197] All steps in this method were performed at 5°C, pH 7.5, unless otherwise stated.
[0198]A 25% by weight suspension of wet cell pulp of A. facilis 72W (ATCC 55746) was prepared with 20 mM Tris pH 7.5, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), and 2.0 mM dithiothreitol.
[0199] Extracts of the suspension were prepared by a French cell press (American Instrument Co., Silver Springs, MD, USA) according to methods known in the art. After centrifugation at 27,500g for 30 minutes to remove cellular debris, a 20-55% ammonium sulfate fraction of the extract was prepared and then concentrated by overnight precipitation after addition of solid ammonium sulfate to 65% saturation. The concentrated protein pellet was dissolved using a minimal volume of 20 mM Tris pH 7.5 (buffer A) and desalted over a PD10 column containing Sephadex G-25 resin (Pharmacia).
[0200] After desalting, the concentrated protein extract was separated by anion exc...
Embodiment 2
[0202] Isolation of DNA fragments showing high homology to known nitrilases
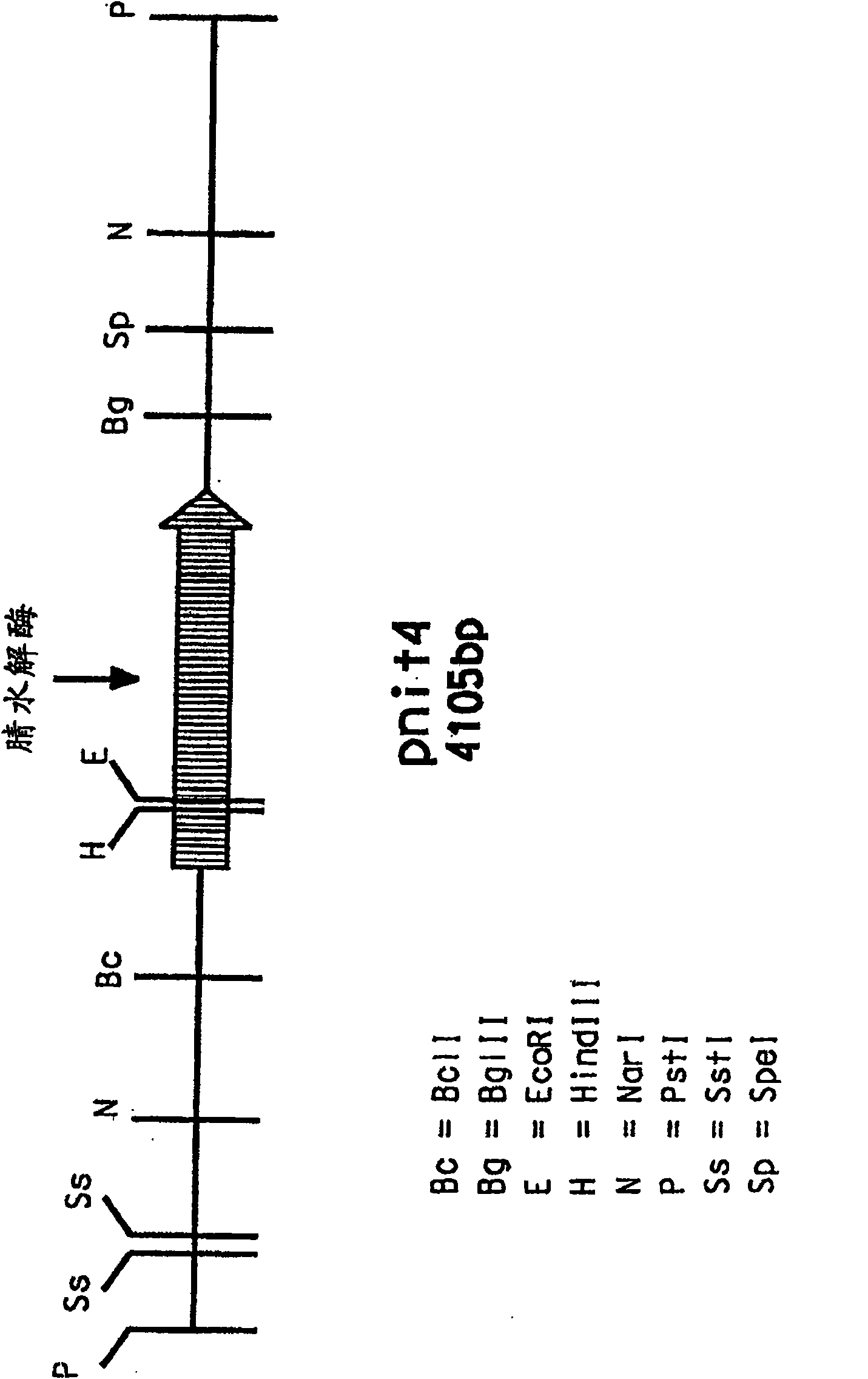
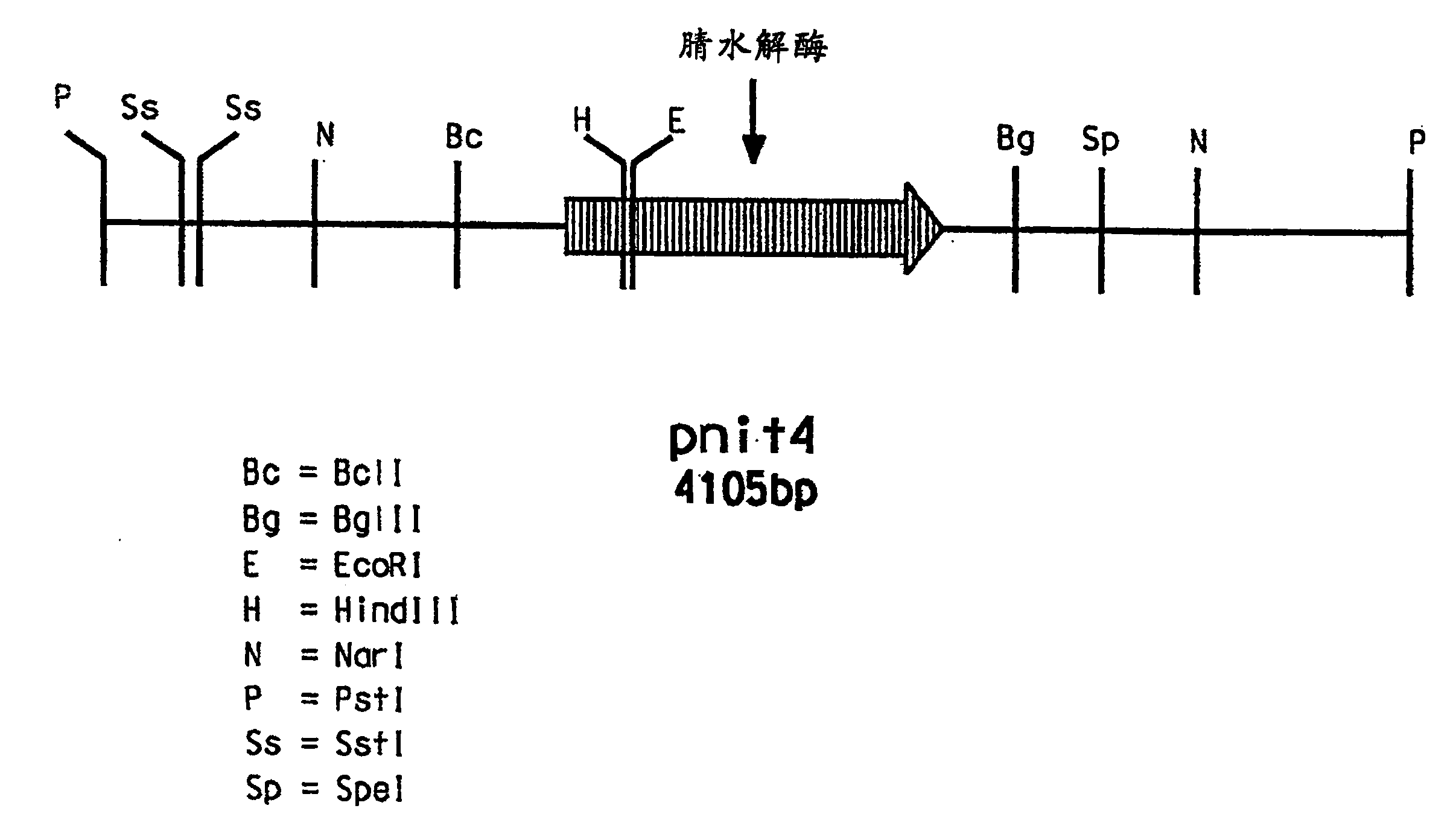
[0203] Acidovorax facilis 72W (ATCC 55746) genomic DNA was isolated by a modified standard method (Maniatis) adding two rounds of phenol-chloroform extraction. The DNA thus isolated was used as a target for PCR amplification using primers 1F (SEQ ID NO: 1) and 7R (SEQ ID NO: 2). The obtained amplified products were cloned into pGem-T vector (Promega, Madison, WI), and sequenced using methods commonly used in the art. The identity of the 385bp DNA fragment (SEQ ID NO: 3) produced by cloning pJJ28-5 with the DNA sequence of Rhodococcus rhodochrous K22 nitrilase (GenBank accession number D12583) was 74.1%.
Embodiment 3
[0205] Locate the chromosomal segment containing a sequence highly homologous to a 385bp partial gene segment
[0206] 1-3 μg of high molecular weight genomic DNA samples of A. facilis 72W were digested with the restriction enzyme Pstl in buffer provided by the manufacturer (Life Technologies (Gibco-BRL), Gaithersburg, MD, USA). Digested samples were separated on agarose gels and Southern blotted on nylon membranes. A probe containing the 385 bp nitrilase gene fragment (SEQ DI NO: 3) was prepared by cutting pJJ28-5 at the restriction sites flanking the insert. Hybridization was performed at 60°C, followed by high stringency washing (0.1×SSC, 0.1% SDS, 60°C, 15 minutes). Probe labeling, hybridization and dot blot detection and subsequent Southern blotting experiments were performed using the ECL Random Primer Labeling and Detection System Type II (Amersham International plc, Buckinghamshire, England).
[0207] The PstI digest yielded a single 4.1 kb band when hybridized to ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com