Compositions of prokaryotic phenylalanine ammonia-lyase variants and methods of using compositions thereof
a technology of phenylalanine ammonialyase and composition, which is applied in the field of prokaryotic phenylalanine ammonialyase (pal) variants, can solve the problems of limited use of pal and lack of concerted efforts to improve these parameters, and achieves enhanced properties, potent catalytic activity, and effective treatment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Cloning of Nostoc punctiforme and Anabaena variabilis PAL DNA Manipulations
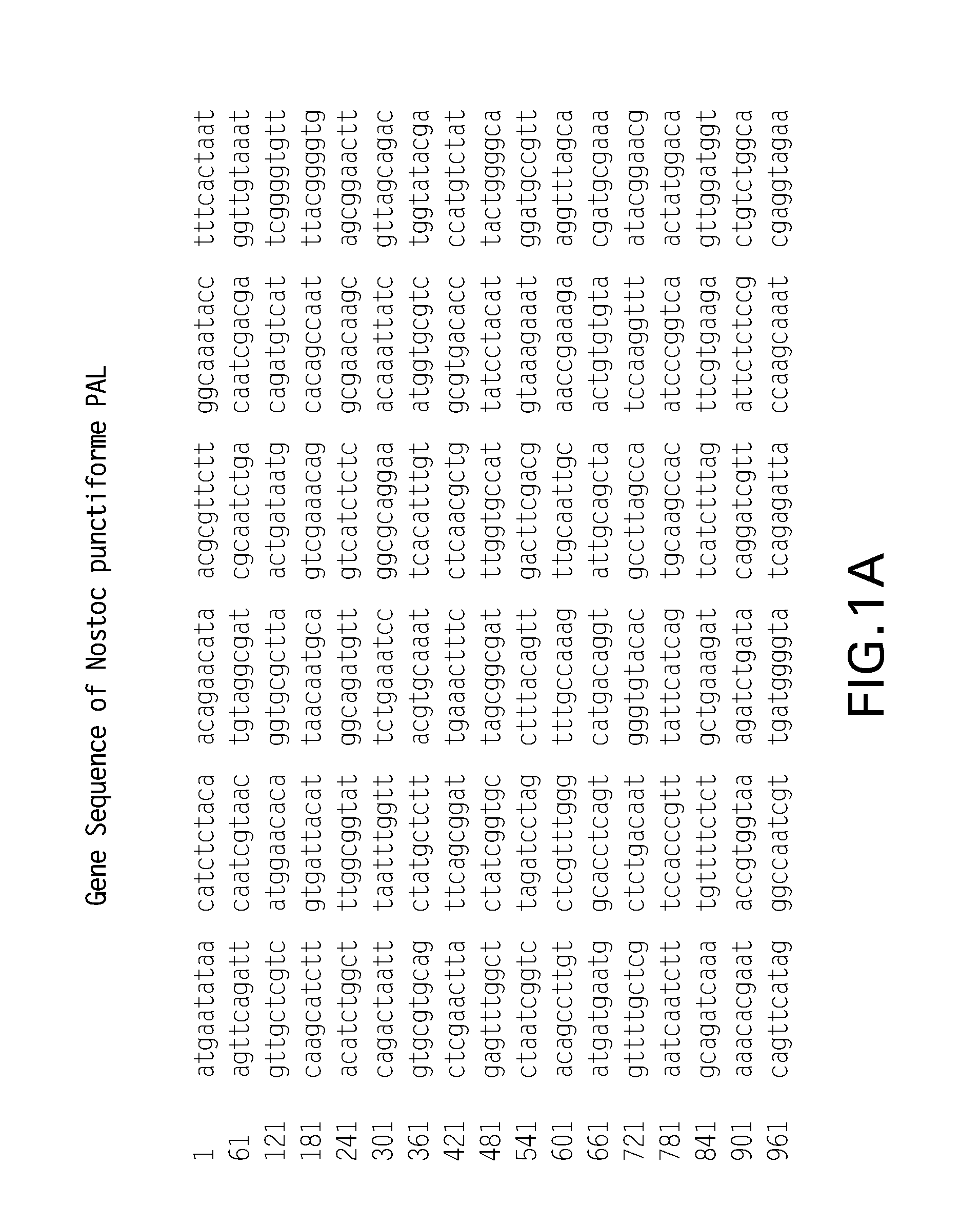
[0221]N. punctiforme genomic DNA was purchased from ATCC (29133D) and the PAL gene (ZP—00105927) was PCR-amplified from primers 5′-CACTGTCATATGAATATAACATCTCTACAACAGAACAT-3′ (SEQ ID NO:12) and 5′-GACAGTGGCGGCCGCTCACGTTGACTTTAAGCTCGAAAAAATATG-3′ (SEQ ID NO:13). The resulting PCR product was digested with NdeI and NotI and the 1.7 kb fragment was ligated into pET-28a(+) and pET-30a(+) (Novagen) for N-His tagged and untagged, respectively.
[0222]A. variabilis cells were purchased from ATCC (29413). Genomic DNA was extracted (Qiagen) and the PAL gene (YP—324488) was amplified by SOE-PCR to remove an NheI site. Primer 1 (5′-CACTGTGCTAGCATGAAGACACTATCTCAAGCACAAAG-3′) (SEQ ID NO:14) and primer 2 (5′-GGAAATTTCCTCCATGATAGCTGGCTTGGTTATCAACATCAATTAGTGG-3′) (SEQ ID NO:15) were used to amplify nucleotides 1-1190 and primer 3 (5′-CCACTAATTGATGTTGATAACCAAGCCAGCTATCATGGAGGAAATTTCC-3′) (SEQ ID NO:16) and primer 4 (5′-CACTGTGCGG...
example 2
Purification of NpPAL and AvPAL
[0227]The cultures were centrifuged in a bench-top centrifuge at 5,000 g for 20 minutes and the supernatant discarded. The cell pellets were typically frozen at −70° C. prior to further processing. Upon thawing, the cell pellets were suspended to approximately 80 optical density units (600 nm) in TBS (25 mM Tris, 150 mM NaCl, pH 7.8). The cells were lysed by two passes through an APV pressure homogenizer at 12-14,000 psi. The crude lysate was then heat-treated at 55° C. for 2 hours. The lysate is centrifuged at 10,000 g for 30 minutes and the supernatant retained and filtered with a 0.2 μm vacuum filter (Corning).
[0228]The PAL was purified from the clarified lysate by passage sequentially over a butyl 650M column (Tosoh BioSciences) and a MacroPrep High Q column (BioRad). The eluted product showed a high level of purity by both SDS PAGE and reverse phase HPLC.
example 3
Generation of Pegylated PAL Variants
[0229]A method for pegylation of PAL from Rhodosporidium toruloides (RtPAL) is described below. Similar methods are used for pegylation of prokaryotic PAL (e.g., NpPAL or AvPAL) are described in EXAMPLE 6.
[0230]Pegylation uses modifications of literature methods (Hershfield, et al., (1991), ibid.; U.S. Pat. No. 6,057,292; Lu, et al., Biochemistry 40(44):13288-13301 (2001); Nektar Therapeutics, 2003 catalog). Activated PEGs include both the linear PEG succinimidyl succinates (mPEG-SPA, MW 5 kDa or MW 20 kDa) and the branched PEG hydrosuccinimides (mPEG2-NHS ester, MW 10 kDa or MW 40 kDa), which are both capped on one end with a methoxy group and available from Nektar Therapeutics; experimental determination of optimal pegylated proteins is normally required (Veronese, et al., J. Bioactive Compatible Polymers 12:196-207 (1997)). Optimal pegylation conditions are determined using different ratios of PAL:PEG (taking into account the ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com