Methods for diagnosing irritable bowel syndrome
a technology for ibs and gastrointestinal disorders, applied in the field of microbiology and gastrointestinal health, can solve the problems of difficult diagnosis of ibs, high rate of absenteeism from work, and significant impairment of quality of life, and achieve the effect of increasing the level of nucleic acids
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Comparison of the Fecal Microbiota of IBS and Healthy Subjects
Study 1
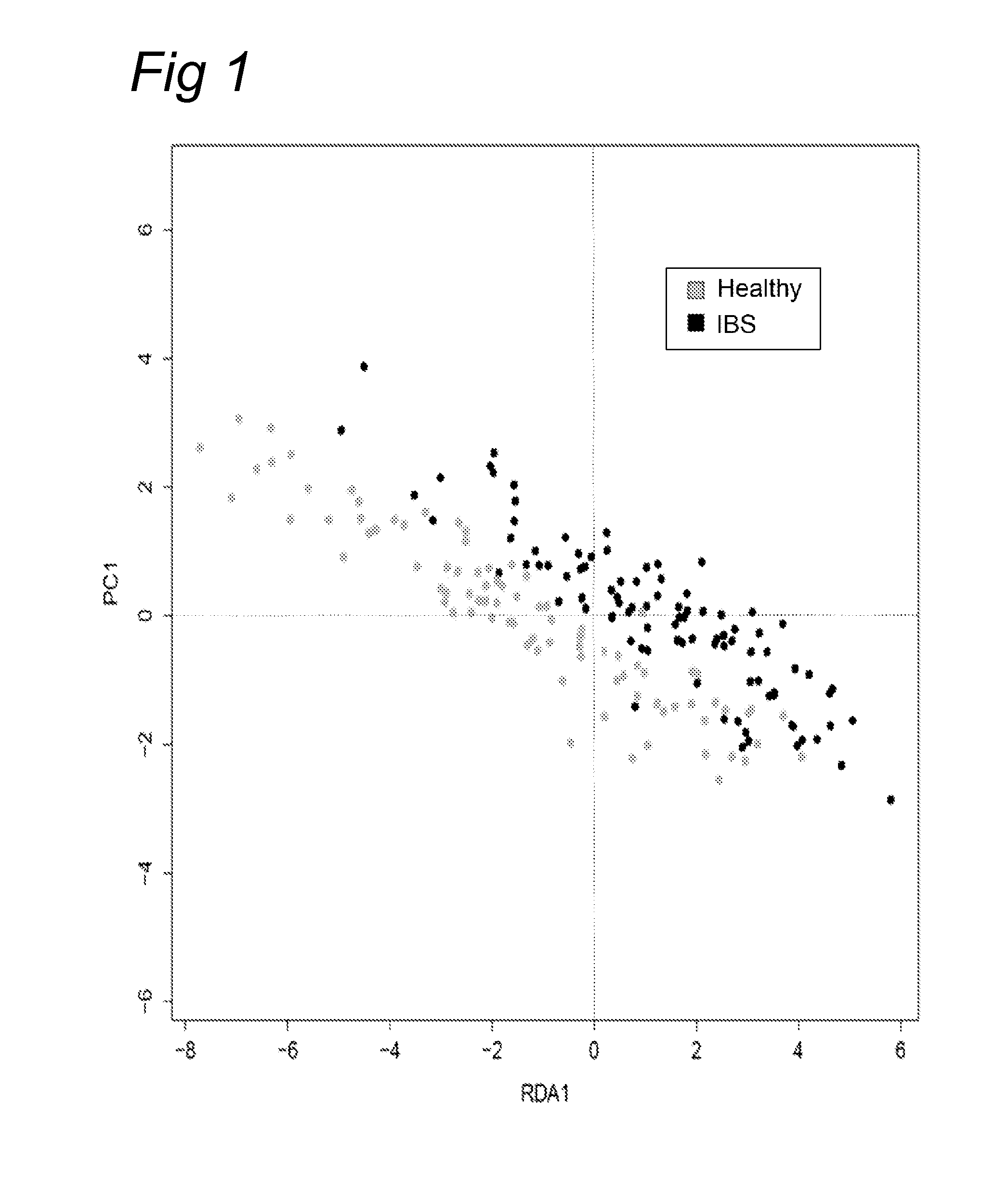
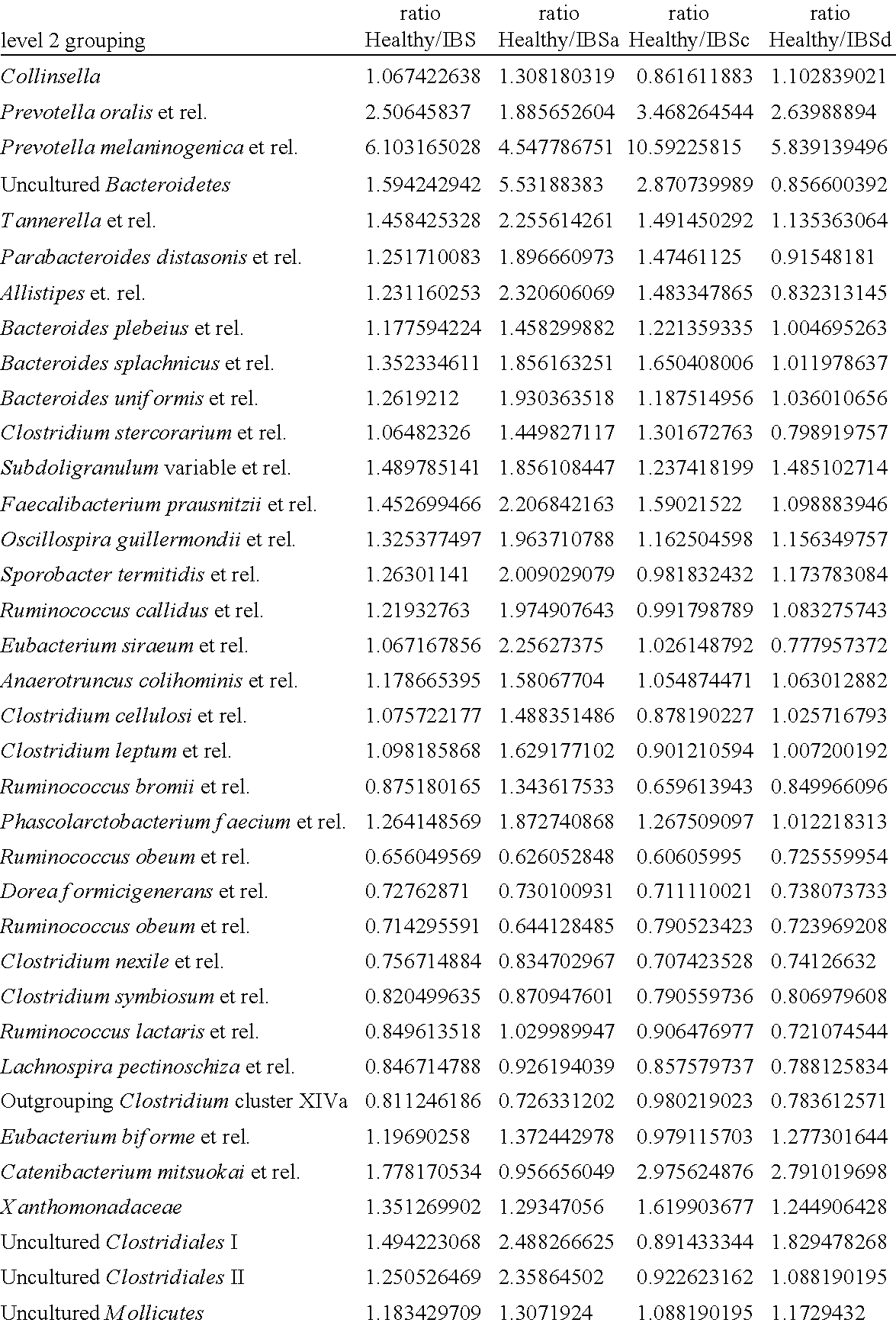
[0104]Fecal samples were obtained from a first study (Study 1) of a total of 62 IBS subjects including 19 with IBS-C, 25 with IBS-D and 18 with IBS-A, and a total of 46 healthy individuals that were age and gender matched. Microbial DNA was isolated from these fecal samples following the method of Ahlroos & Tynkynnen (2009, supra) and used for profiling using the HITChip phylogenetic microarray using 3699 distinct HIT probes as described (Rajilic-Stojanovic et al., 2009, supra). Based on the intensity of the hybridization signals obtained in the HITChip analysis from the 62 IBS subjects and 46 healthy individuals a total of 36 level 2 microbial groups from the total of over 100 groups was found to be reacting significantly different between IBS and healthy subjects (see Table 1 above). The identified microbial groups can be developed as biomarker as described above. Moreover, the differences in microbiota can be co...
example 2
Identification of IBS- and Healthy-Specific Oligonucleotides
[0105]In order to further define the specific oligonucleotide probes that were reacting different in the IBS subjects as compared to the healthy controls, the hybridization of all 3,699 HIT probes of the HITChip in Study 1 (Example 1) was analyzed, resulting in a total of 100 HIT probes were found to be differentially hybridizing (Tables 2 and 4). A total of 34 HIT probes (oligonucleotides having SEQ ID Nos:1-27, 70-71, 73-77, 99-100) showed a significantly higher hybridization signal in the IBS subjects than the healthy individuals, while a total of 66 (oligonucleotides having SEQ ID Nos:28-69, 72, 78-98) showed less hybridization in the IBS subjects than the healthy subjects, respectively. The sequences of these oligonucleotides are disclosed in Tables 2 and 4 and allow the development of specific probes as described above. Moreover, these probes can be used to screen the 16S rDNA databases for complete 16S rRNA sequences...
example 3
Further Analysis of the Differences in Fecal Microbiota of IBS and Healthy Subjects
[0106]To further substantiate the differentiation of IBS subjects and healthy controls based on fecal microbiota, a second set of samples was analyzed that included a total of 33 IBS subjects that were not further differentiated and 43 healthy controls that were age and gender matched (Study 2). Fecal samples were obtained from these 77 individuals and microbial DNA was isolated from these following the repeated bead beating method as described (Yu & Morrison, 2004, supra). This DNA was used for profiling using the HITChip phylogenetic microarray using 3699 distinct HIT probes as described (Rajilic-Stojanovic et al., 2009, supra). As the DNA extraction method differed between Study 1 (Example 1) and Study 2 (the results presented here) as an enzymatic and mechanical lysis method was used, respectively, it was of interest to see the differentiation of the datasets obtained from the HITChip analysis in ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Electrical conductance | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com