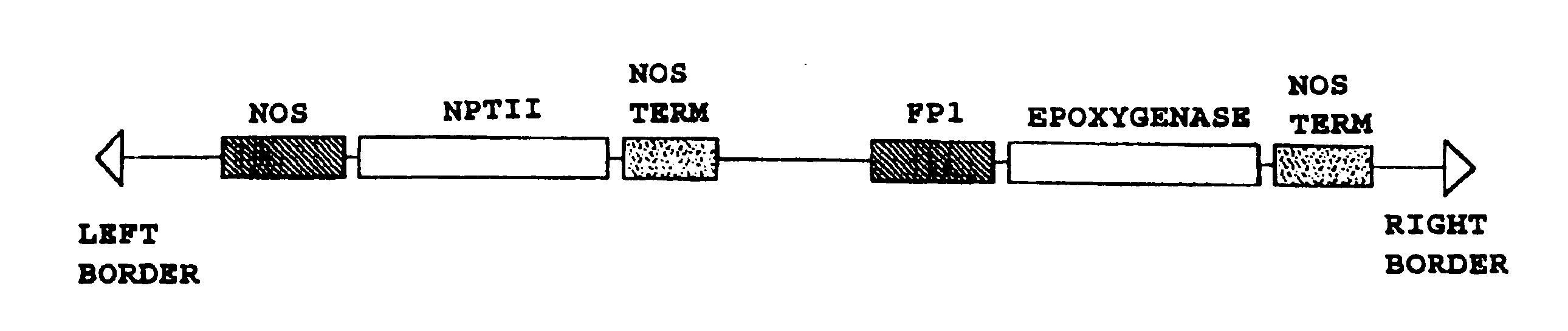
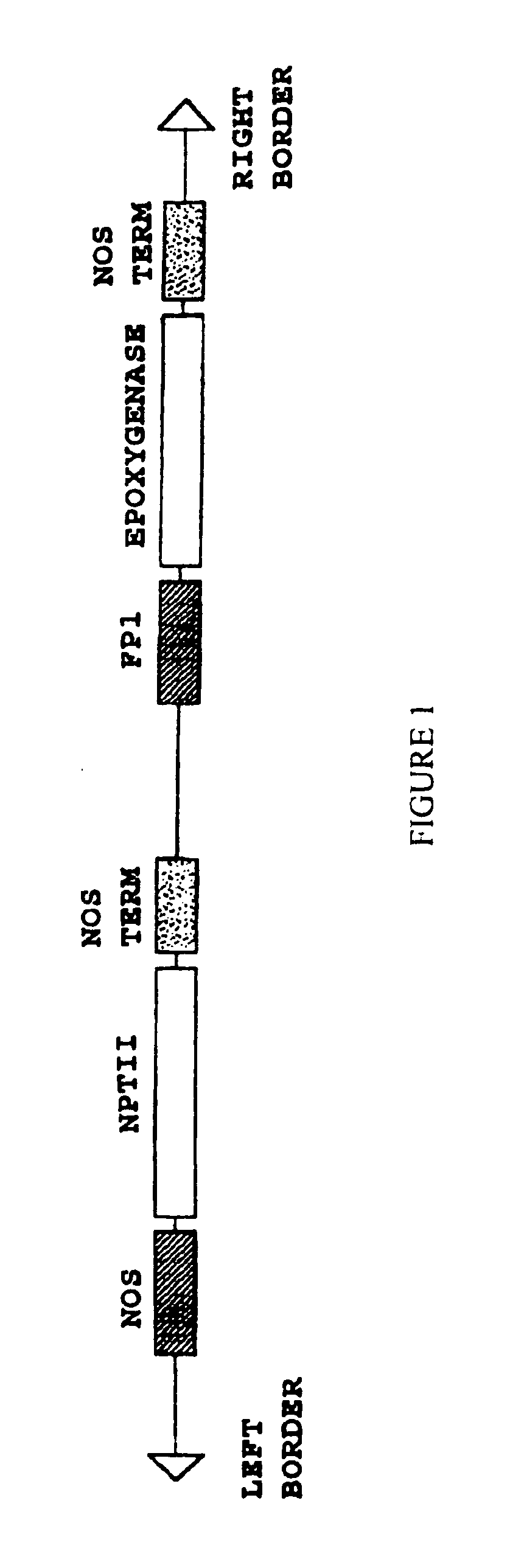
Plant seed comprising vernolic acid
a technology of vernolic acid and vernolic acid, which is applied in the field of new genetic sequences that encode the enzymes of fatty acid epoxygenase, can solve the problems of limiting the commercial utility affecting the development of genetically engineered organisms which produce these fatty acids, and affecting the commercialization of traditional plant breeding and cultivation approaches
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0240] Characterization of epoxy fatty acids in Euphorbia lagascae and Crepis spp. Seed from the wild species Euphorbia lagascae and from various Crepis species were screened by gas liquid chromatography for the presence of epoxy fatty acids. As shown in Table 3, Euphorbia lagascae contains very high levels of the epoxy fatty acid vernolic acid in its seed oil. Seeds from Crepis palaestina were shown to contain 61.4 weight % of vernolic acid and 0.71 weight % of the acetylenic fatty acid crepenynic acid of total fatty acids (Table 3).
TABLE 3Fatty acid composition of lipids derived from seeds ofCrepis alpina, Crepis palaestina and Euphorbia lagascaeRelative distribution (weight %)aFatty acidCrepis alpinaCrepis palaestinaEuphorbiaPalmitic3.95.14.3Stearic1.32.31.8Oleic1.86.322.0Linoleic14.023.010.0Crepyninic75.00.70Vernolic061.458.0Other4.01.23.9
aCalculated from the area % of total integrated peak areas in gas liquid chromatographic determination of methyl ester derivatives of the se...
example 2
Biochemical Characterization of Linoleate Δ12-epoxygenases in Euphorbia lagascae and Crepis palaestina
[0241] The enzyme, linoleate Δ12-epoxygenase synthesizes vernolic acid from linoleic acid. Linoleate Δ12-epoxygenases derived from Euphorbia lagascae and Crepis palaestina are localized in the microsome. The enzymes from these species at least can remain active in membrane (microsome) fractions prepared from developing seeds.
[0242] Preparations of membranes from Euphorbia lagascae and assays of their epoxygenase activities were performed as described by Bafor et al. (1993) with incubations containing NADPH, unless otherwise indicated in Table 4. Lipid extraction, separation and methylation as well as GLC and radio-GLC separations were performed essentially as described by Kohn et al. (1994) and Bafor et al. (1993).
[0243] Preparations of membranes from Crepis alpina and Crepis palaestina were obtained as follows. Crepis alpina and Crepis palaestina plants were grown in green house...
example 3
Strategy for Cloning Crepis palaestina Epoxygenase Genes
[0249] Cloning of the Crepis palaestina epoxygenase genes relied on the characteristics of the C. palaestina and C. alpina enzymes described in the preceding Examples.
[0250] In particular, poly (A)+RNA was isolated from developing seeds of Crepis palaestina using a QuickPrep Micro mRNA purification kit (Pharmacia Biotechnology) and used to synthesize an oligosaccharide d(T)-primed double stranded cDNA. The double stranded cDNA was ligated to EcoRI / NotI adaptors (Pharmacia Biotechnology) and a cDNA library was constructed using the ZAP-cDNA Gigapack cloning kit (Stratagene).
[0251] Single-stranded cDNA was prepared from RNA derived from the developing seeds of Crepis alpina, using standard procedures. A PCR fragment, designated as D12V (SEQ ID NO: 7), was obtained by amplifying the single-stranded cDNA using primers derived from the deduced amino acid sequences of plant mixed-function monooxygenases.
[0252] The D12V fragment w...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com