Method for attachment of silylated molecules to glass surfaces
a silylated molecule and glass surface technology, applied in the field of microarray biomolecule detection technology, can solve the problems of inability to give reproducible silane, and inability to achieve reproducible optimum surface loading, so as to enhance the detection of target analytes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Preparation of DNA Array Chips
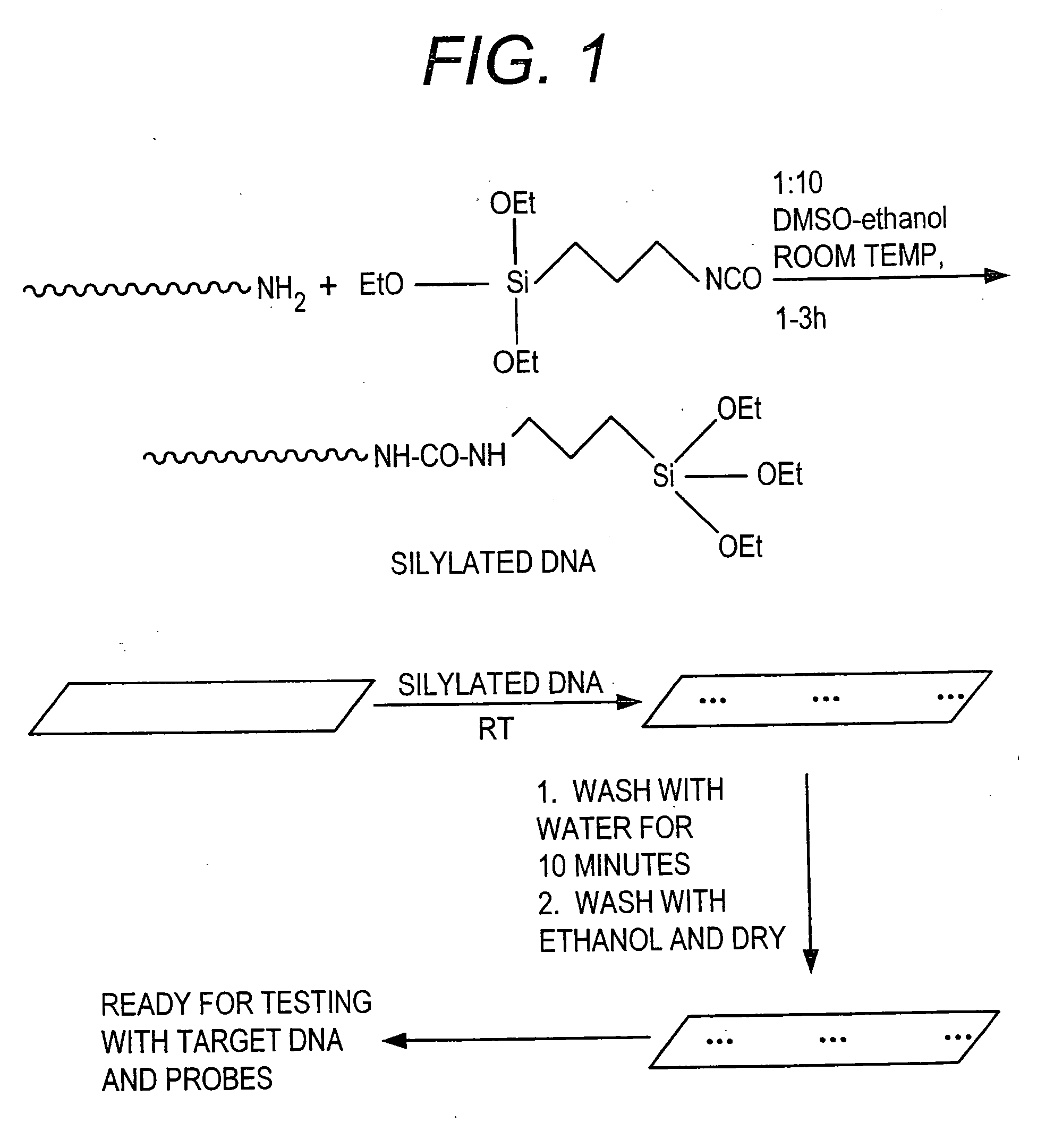
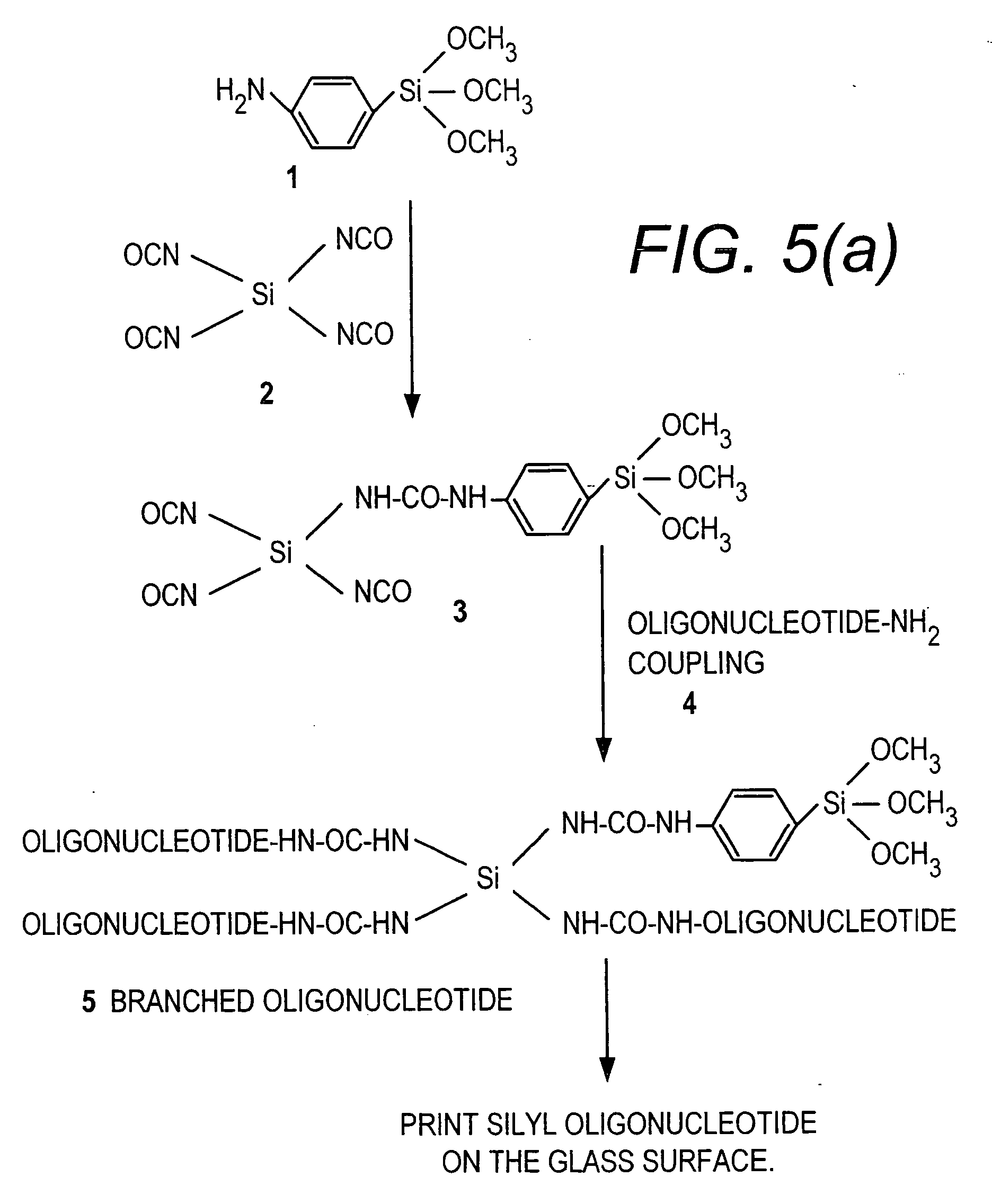
[0058] This Example provides a general procedure for the covalent attachment of a molecule, e.g., 3′ or 5′-silylated DNA, directly to surfaces such as pre-cleaned glass surface via single silylated molecule or dendritic silylated molecule procedure.
(a) Method No. 1
[0059] As shown in FIG. 1, a method is shown for attaching a 3′-amino or 5′-amino DNA molecule to a pre-cleaned glass surface. 3′-Amine linked DNA is synthesized by following standard protocol for DNA synthesis on DNA synthesizer. The 3′ amine modified DNA synthesized on the solid support was attached through succinyl linker to the solid support. After synthesis, DNA attached to the solid support was released by using aqueous ammonia, resulting in the generation of a DNA strand containing a free amine at the 3′-end. The crude material was purified on HPLC, using triethyl ammonium acetate (TEAA) buffer and acetonitrile. The dimethoxytrityl (DMT) group was removed on the column itself using ...
example 2
Detection of Factor V Target Sequence Using a DNA Array Chip
[0063] This Example illustrates that DNA plates prepared as described in Example 1 are useful for sandwich hybridization assays for detection of nucleic acid targets.
(a) Gold Colloid Preparation:
[0064] Gold colloids (13 nm diameter) were prepared by reduction of HAuCl4 with citrate as described in Frens, Nature Phys. Sci., 241, 20 (1973) and Grabar, Anal. Chem., 67, 735 (1995). Briefly, all glassware was cleaned in aqua regia (3 parts HCl, 1 part HNO3), rinsed with Nanopure H2O, then oven dried prior to use. HAuCl4 and sodium citrate were purchased from Aldrich Chemical Company. Aqueous HAuCl4 (1 mM, 500 mL) was brought to reflux while stirring. Then, 38.8 mM sodium citrate (50 mL) was added quickly. The solution color changed from pale yellow to burgundy, and refluxing was continued for 15 min. After cooling to room temperature, the red solution was filtered through a Micron Separations Inc. 1 micron filter. Au colloid...
example 3
Detection of M13 Target Sequence Using DNA Array Chip
[0078] In this Example, probe was targeted directly to the capture strand and a detection assay was performed. Plates Nos. 1-3 were prepared as described in Example 1 (method no. 1). In Plates 2 & 3, probes (FIG. 6) were clearly hybridized to the capture strand within 45 minutes. The gold colloid nanoparticles hybridized to the capture were clearly visible before silver amplification. In plate no 1 (FIG. 6), a different probe was used and the assay was developed to show the specificity. After silver stain development, signals were not shown on the glass surface even after silver amplification. This experiment established the specificity of the DNA chip prepared in accordance with the invention.
[0079] M13 Capture sequence:
(SEQ ID NO: 8)5′-tga aat tgt tat c-NH-CO-NH--Si-(OEt)3-3′
[0080] Probe used on plates Nos. 2-3 plates:
3′-act tta aca ata g-a20-Epi-5′(SEQ ID NO: 9)
[0081] On plate no.1, a detection probe 3′-t taa cac tcg c-a2...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com