Real-time polymerase chain reaction-based genotyping assay for single nucleotide polymorphism
a polymorphism and genotyping technology, applied in biochemistry, sugar derivatives, organic chemistry, etc., can solve the problems of allelic discrimination and insufficient single base pair difference at the 3′ end of the primer, and achieve the effect of preventing the amplification of non-matching primers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
MDR1 Single Nucleotide Polymorphisms Genotyping
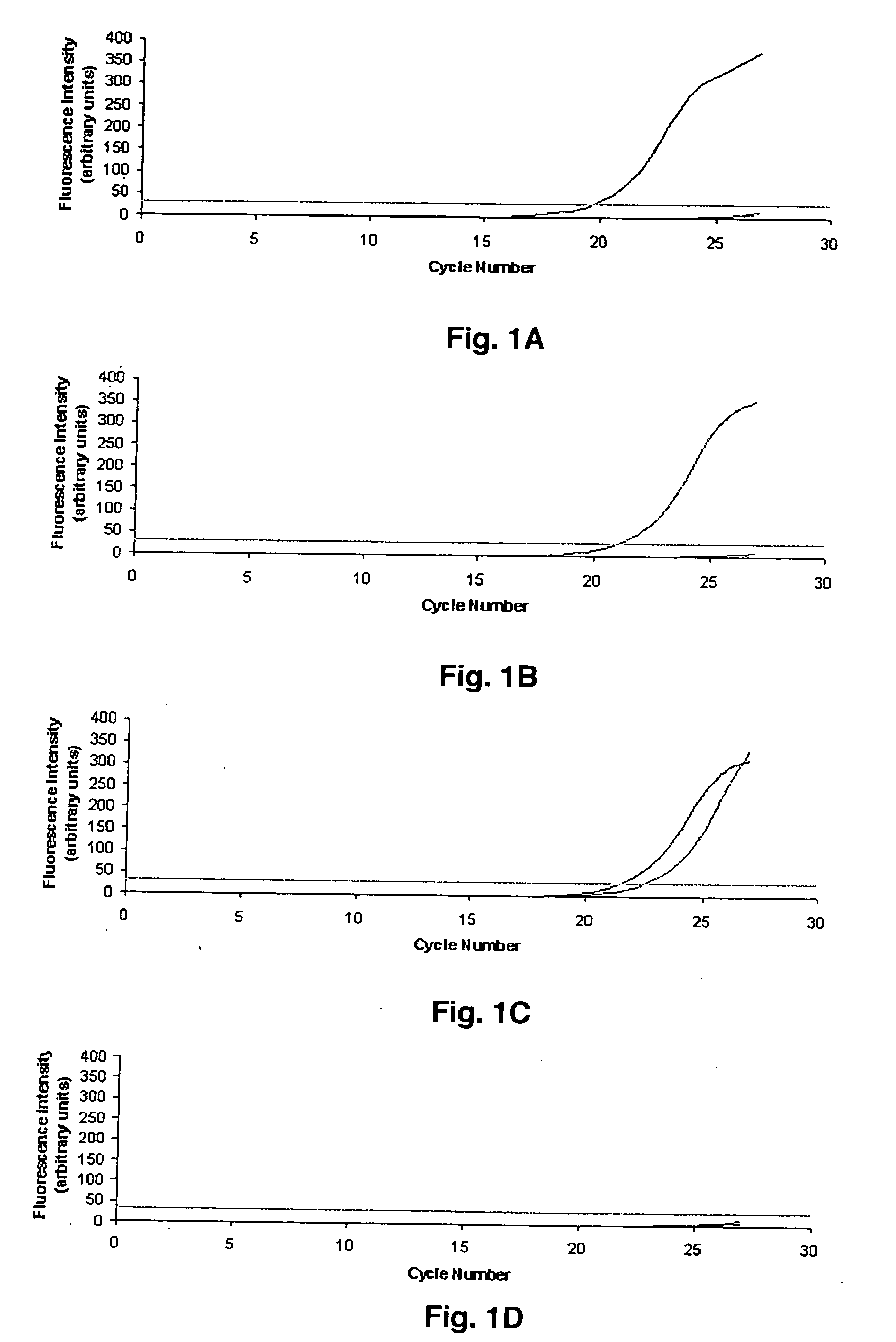
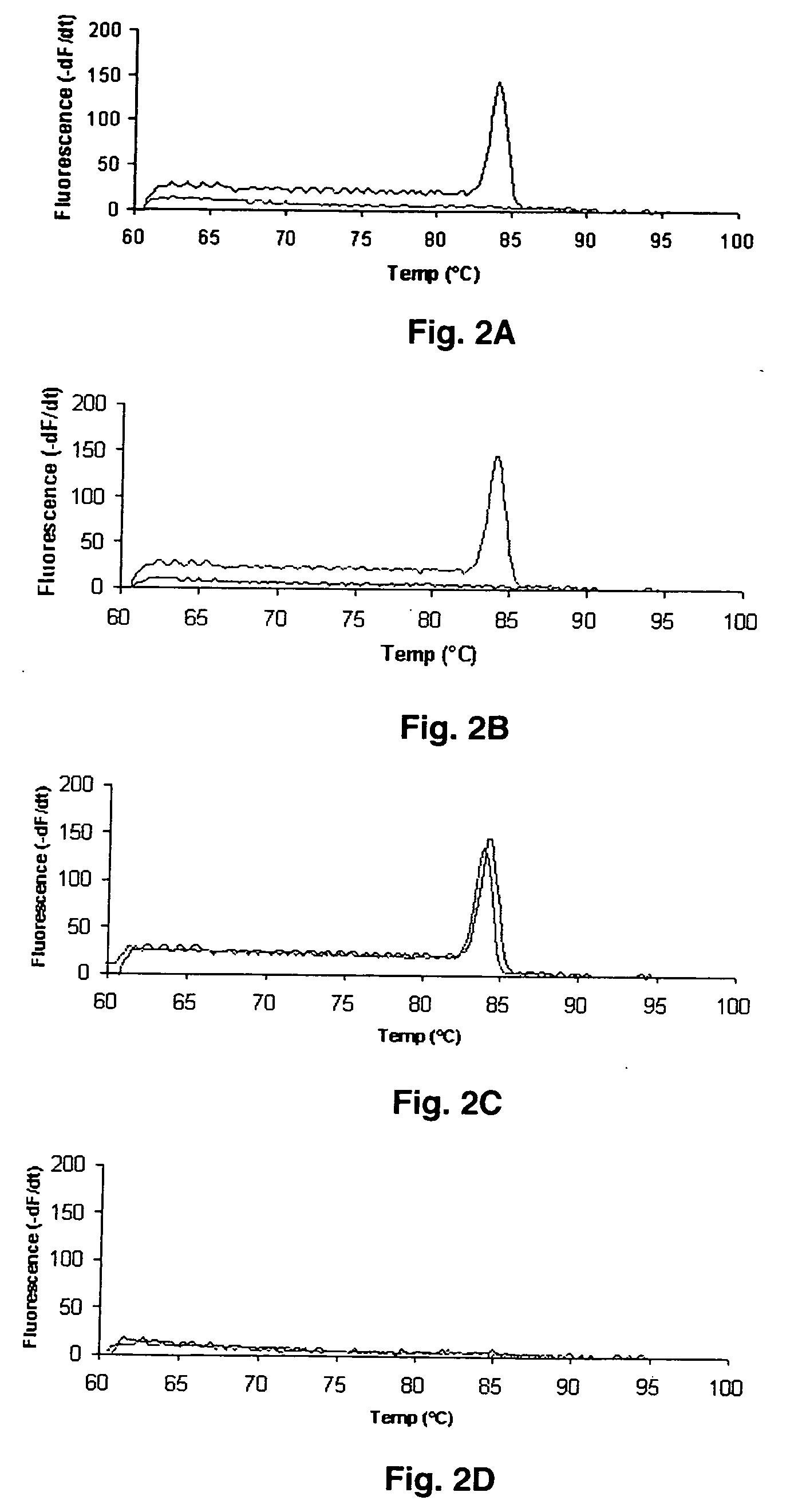
[0029] The present example describes a real-time PCR assays for the rapid detection of the MDR1 single nucleotide polymorphisms C3435T and G2677T. These methods can be readily applied to investigate the effect of MDR1 polymorphic expression on pharmacokinetic and pharmacodynamic variability of P-gp substrates.
example 2
Primer Design
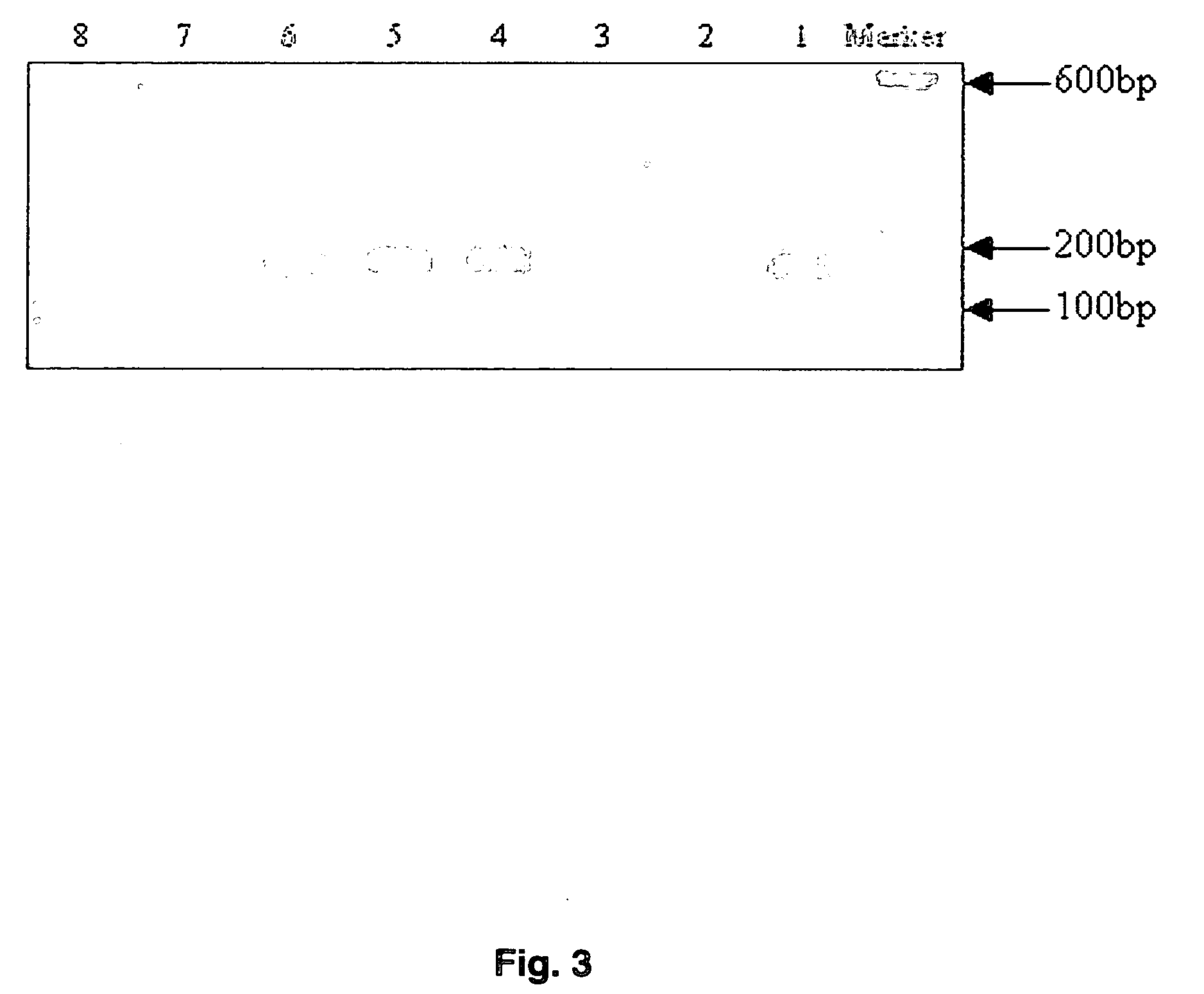
[0030] PCR primers are listed in Table 1. Oligonucleotide primers were designed based on the published MDR1 sequence (Genbank #AC005068) using the online program Primer3. Hairpin structures and primer-dimers were predicted with Oligo Toolkit™. The primers were synthesized by Integrated DNA Technologies (Coralville, Iowa). Expected amplicon lengths were 216 base pairs (bp) and 134 bp for G2677T and C3435T, respectively. Discrimination between wild type and mutant alleles was achieved using PCR amplification of specific alleles modified to prevent non-Watson Crick base pairing (Okimoto & Dodgson, 1996; Sommer et al., 1992; Bottema et al., 1993; Newton et al., 1989). Briefly, the first nucleotide difference (C or T) between sense primers (3435W and 3435M) used to discriminate between wild type and mutant 3435 alleles is located at the 3′ terminal base. The second primer base change (A to G) located 3 bases from the 3′ end generates an internal primer / template mismatch, a...
example 3
Real-Time PCR Amplification
[0031] The Smart Cycler (Cepheid, Sunnyvale, Calif.) was used to monitor PCR amplification using SYBR™ Green I (Molecular Probes, Eugene, Oreg.), a nonspecific double-stranded DNA intercalating fluorescent dye. Thus, to achieve allelic discrimination between wild type and mutant alleles, two physically separate PCR reactions containing either wild type or mutant-specific primers were performed. All reactions were carried out in a total volume of 25 μL. Reaction conditions were identical for G2677T and C3435T except where noted. Each reaction mixture contained a 1:12,500 dilution of SYBR™ Green I nucleic acid gel stain 10,000× in dimethyl sulfoxide (DMSO) (Molecular Probes); 0.2 mM of dATP, dCTP, dGTP, and dTTP; 200 nM of both forward and reverse primers; 1.0 U of Taq DNA polymerase (Promega, Madison, Wis.); 6% DMSO; and 20 to 120 ng of genomic DNA in 1×PCR buffer (pH 8.3, 10× solution containing 100 mM Tris-HCl, 500 mM KCl, 15 mM MgCl2 and 0.01% gelatin)...
PUM
| Property | Measurement | Unit |
|---|---|---|
| total volume | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com