Pichia pastoris omega3-fatty acid dehydrogenase promoter sequence and use thereof
A fatty acid dehydrogenase and promoter sequence technology, applied in the biological field, can solve the problems of promoter sequence cloning and analysis that have not yet been reported, and achieve the effect of major application prospects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Example 1. Isolation of Pichia pastoris ω3-fatty acid dehydrogenase gene promoter sequence (ppw3P)
[0061] The total DNA of Pichia pastoris was extracted, and the genome was digested with SalI. Using the fragment (663bp-1090bp, SEQ ID NO: 2) obtained by double digestion (BamHI&SalI) of the open reading frame of Mortierella isabellina (Mortierellaisabellina) Δ6-fatty acid dehydrogenase as a linker, the genome and linker after enzyme digestion were molar ratio 1 :3 ratio for overnight ligation. Design and synthesize the following three primers:
[0062] Primer 1: GAACTCCTCAGCGGTGTATGGAACAAAGAC
[0063] Primer 2: ATGACCTGTT GCCTTATGAT GC
[0064] Primer 3: GGCGATTGT GTTCTCGCTC
[0065] Using the ligation product as a template, perform a PCR reaction:
[0066] Reaction component Addition amount
[0067] Buffer (10×)
[0068] (Containing 20mmol / L MgCl 2 ) 5μl
[0069] dNTP (2.5mmol / L) 2μl
[0070] Primer 1 (1μmol / L) 2μl
[0071] Primer 2 (1μmol / L) 2μl
[0072] T...
Embodiment 2
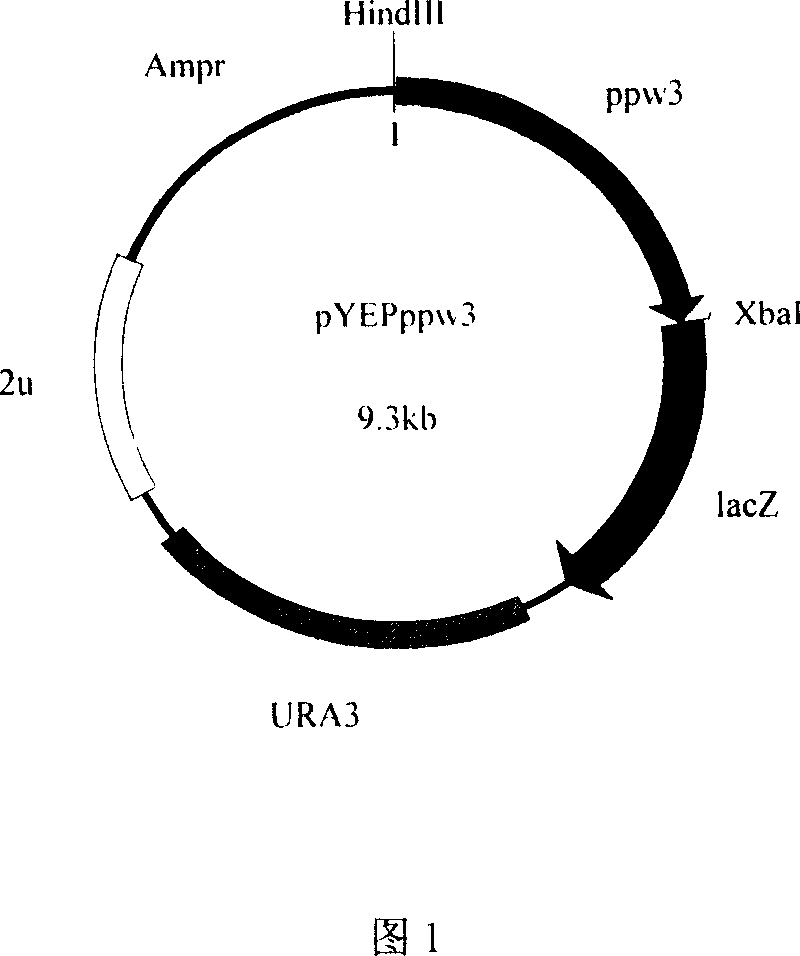
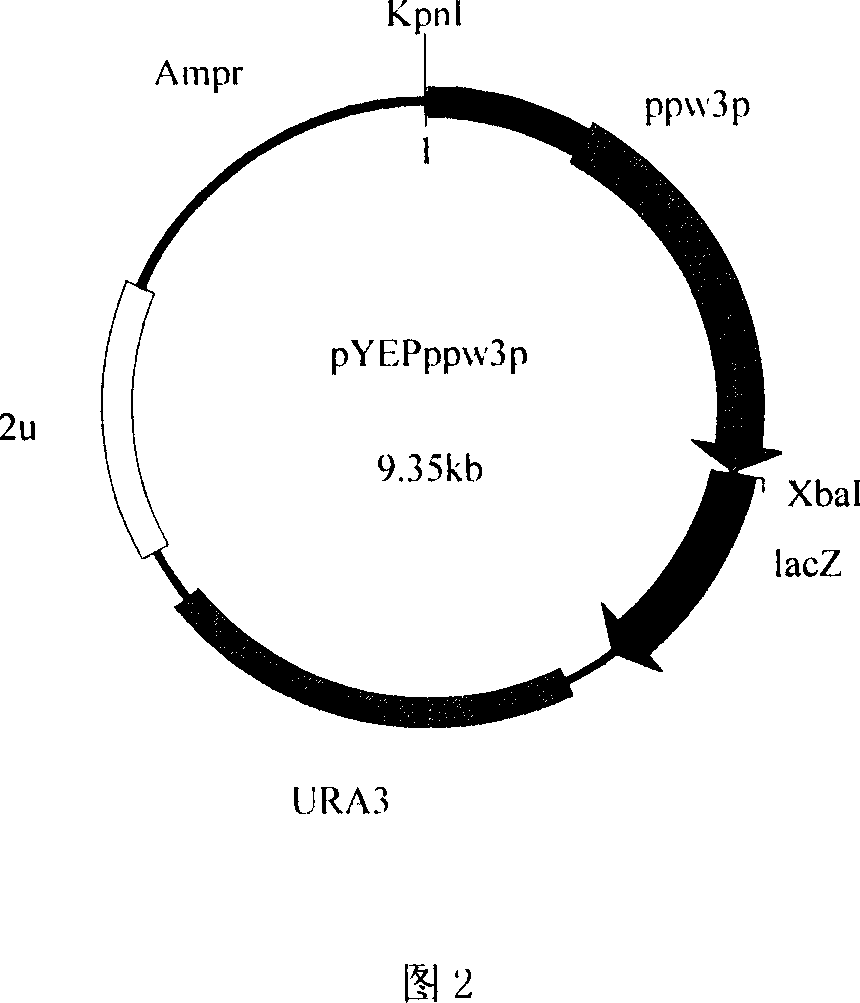
[0078] Embodiment 2: Construction of Saccharomyces cerevisiae recombinant expression vector
[0079] According to the sequence shown in SEQ ID NO: 1, 392-779, gene-specific amplification primers (primers 5 and 6) were designed to isolate its potential promoter sequence:
[0080] Primer 4: 5-GGCAAGCTTATGTCAAAAGTCACTGTTTCGGG-3`;
[0081] Primer 5: 5`GCCTCTAGATTAGGTATCCTTAGG-3`;
[0082] Primer 6: 5`-GGCGGTACCGCCGAAGAAAGTAGAAGAGAAG-3`
[0083] The 5' ends of primers 4 and 5 contain HindIII and XbaI restriction sites respectively, which are used to separate ω 3 - Open reading frame (ppw3) of fatty acid dehydrogenase. The 5' ends of primers 6 and 5 contain KpnI and XbaI restriction sites respectively, which are used to separate ω 3 - Open reading frame of fatty acid dehydrogenase and its potential promoter sequence (ppw3p). The amplification conditions and reaction components used were the same as above, and then 50 μl of the PCR product and 1 μl of pYEP356 were respectively s...
Embodiment 3
[0084] Example 3: Electric shock-mediated transformation of recombinant plasmid Saccharomyces cerevisiae
[0085] Saccharomyces cerevisiae strain INVSc1 was streak-inoculated from a glycerol tube on a YEPD plate, and cultured at 30°C for 48 hours for activation; a single colony was picked and cultured in 5ml YEPD liquid medium, shaken at 30°C and 250rpm overnight; the OD of the bacterial liquid was detected 600 value, take an appropriate amount of bacterial solution and dilute it in 50ml YEPD liquid medium to make the OD 600 =0.4, continue to culture for 2-4h: centrifuge the cells at 2500rpm, remove the supernatant, add 50ml ice-bathed sterile water to wash twice, and ice-bath 1mol / L sorbitol to wash twice; 2ml 1mol / L sorbitol Resuspend the cells in alcohol and aliquot them in 100 μl / tube, and use them on ice as soon as possible. Add 10 μg of recombinant expression plasmid to 100 μl of competent yeast cells and mix gently; set the MicroPulser to the “ScaI” block, the paramete...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com