Salt algae NADP glyceral dehyde-3-phosdehydrogenase gene clone and protein expression method
A phosphate dehydrogenase and protein expression technology, applied in the field of salina salina NADP-glyceraldehyde-3-phosphate dehydrogenase gene cloning and protein expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1: Cloning of the key enzyme NADP-glyceraldehyde-3-phosphate dehydrogenase gene in the photosynthetic metabolic pathway of Salina
[0027] First, a pair of primers (CRGAPAF: (SEQ ID NO: 1); CRGAPAR (SEQ ID NO: 2)) were synthesized based on the conserved sequence of Chlamydomonas W80 NADP-glyceraldehyde-3-phosphate dehydrogenase gene cDNA (AB035312), A cDNA fragment (176bp) (SEQ ID NO: 3) with the expected size was amplified from the total RNA of Chlamydomonas CC2677 by RT-PCR. Sequence analysis was performed on the obtained cDNA fragment, and it was found that the homology with the corresponding sequence of AB035312 was 89.2%, while the homology with the corresponding amino acid sequence (SEQ ID NO: 4) was 93.1%, and only 4 amino acid sequences were different. We named this fragment CRGAPAf. Then, the Chlamydomonas 2677 glyceraldehyde-3-phosphate dehydrogenase gene fragment CRGAPAf (SEQ ID NO: 3) obtained above was probe-labeled with DIG, and the cDNA library (...
Embodiment 2
[0028]Example 2: Sequence analysis of the key enzyme NADP-glyceraldehyde-3-phosphate dehydrogenase gene in the photosynthetic metabolic pathway of Salina salina: the full-length cDNA sequence of DvGapA1 encodes a protein sequence of 376 amino acids. According to the sequence deduction, its molecular weight is 40.5kDa, and its isoelectric point is 9.05 basic protein. BlastP comparison of the deduced protein sequence found that it has high homology with the protein encoded by the glyceraldehyde-3-phosphate dehydrogenase gene located on the chloroplast from different species, among which the glycerol of Chlamydomonas The amino acid sequence homology of aldehyde 3-phosphate dehydrogenase (BAA94304) is as high as 72%. Through online chloroplast transit peptide prediction ( http: / / www.cbs.dtu.dk / services / ChloroP / ) found that the first 35 amino acids of the protein encoded by the gene we isolated were possible transit peptides, which was consistent with the distribution of chlorop...
Embodiment 4
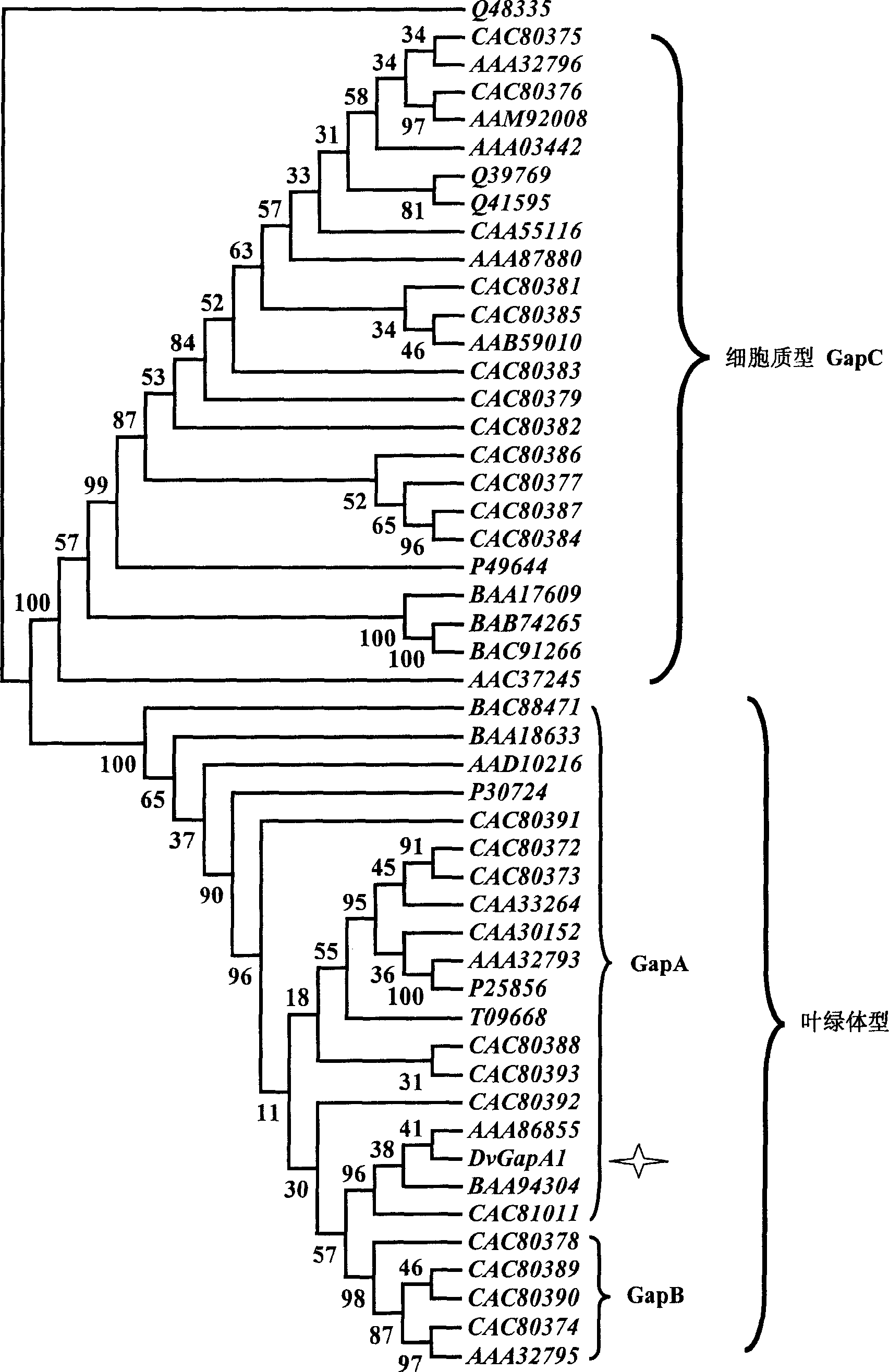
[0029] Example 4: Comparison of protein sequences of glyceraldehyde-3-phosphate dehydrogenase (GapA, GapB) in different species
[0030] Sequence comparison of chloroplast-type glyceraldehyde-3-phosphate dehydrogenases from spinach, Arabidopsis, pea and salina shows that the protein sequence of GapB type has a significantly longer C-terminus than the protein sequence of GapA type , and the homology of this C-terminal sequence from different species is relatively high. The glyceraldehyde-3-phosphate dehydrogenase DvGapA1 of Salina salina obviously belongs to the GapA type and contains conserved amino acid residues in the NADP-binding and enzyme protein catalytic domains, see figure 1 , which shows that salina glyceraldehyde-3-phosphate dehydrogenase may be conserved in the higher order structure.
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com