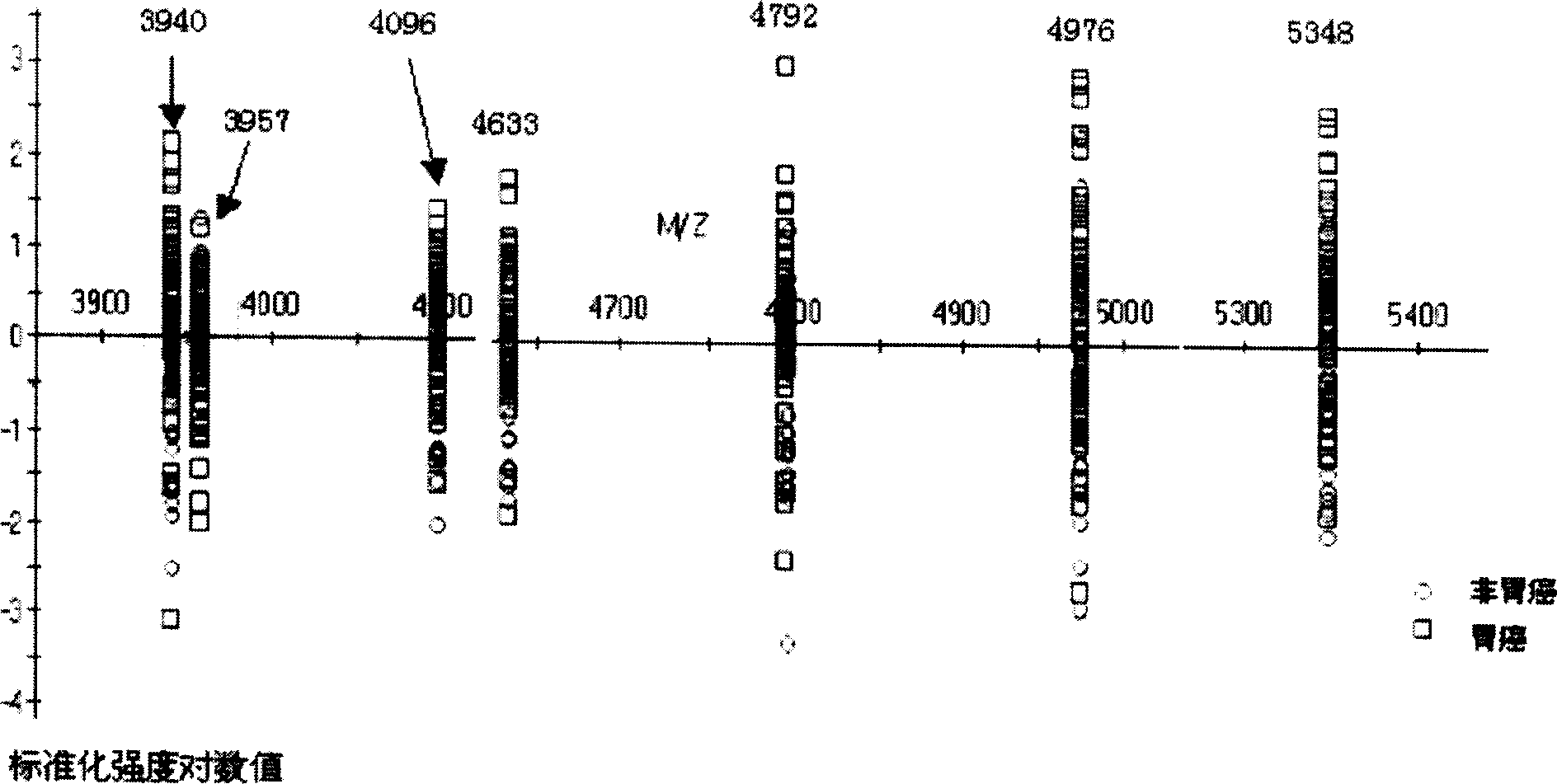
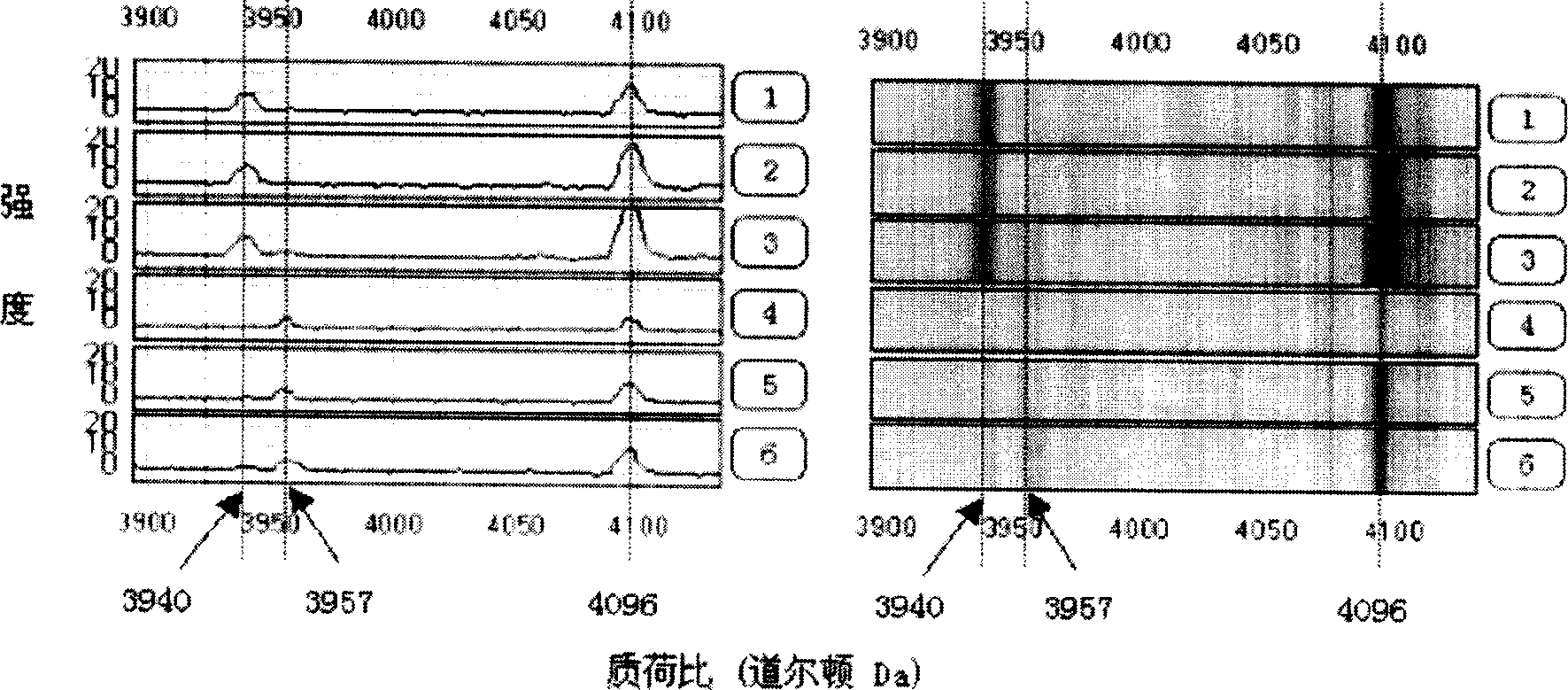
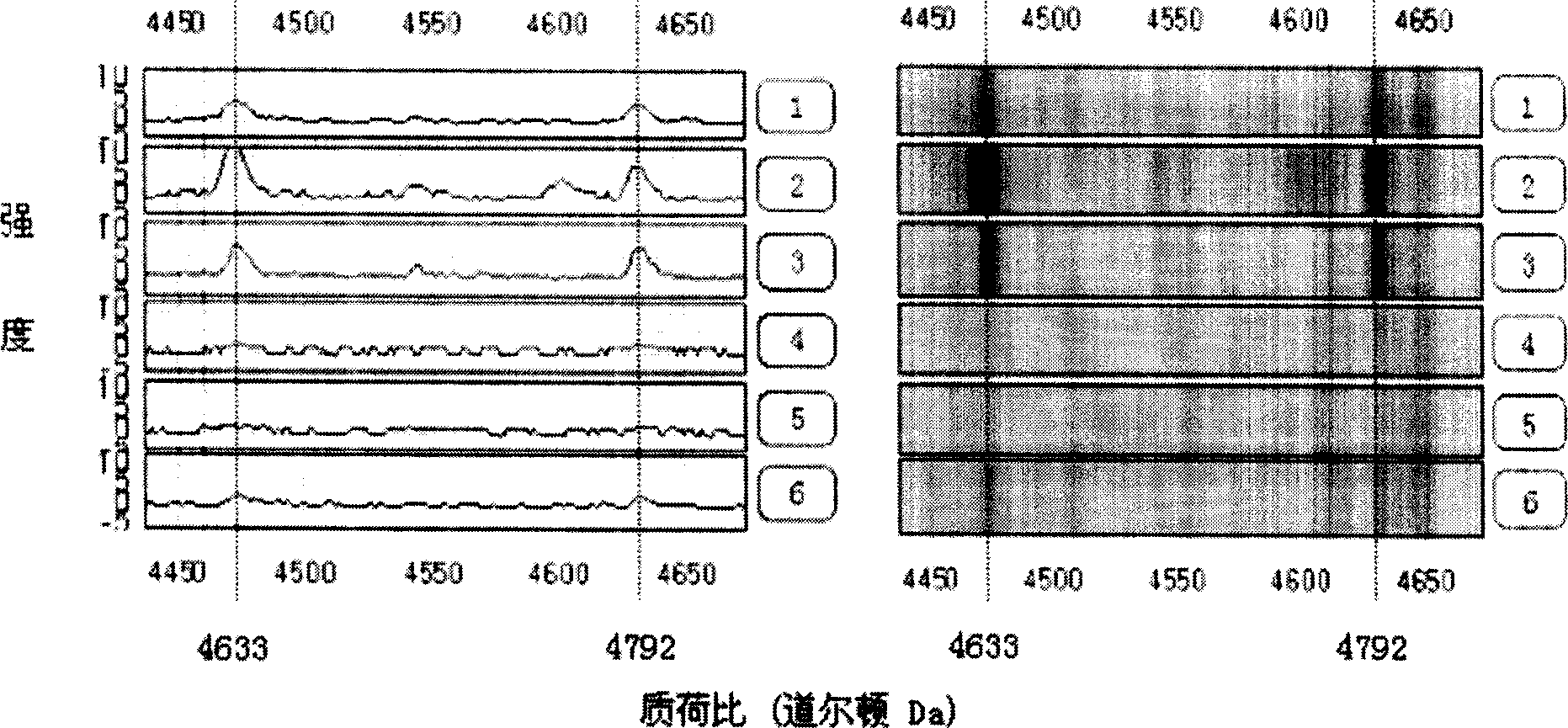
Method for early detecting gastric cancer from blood serum
An early detection, serum technology, applied in the field of early detection of malignant tumors, can solve the problems of electrophoretic separation and detection of insoluble proteins
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] 1. Serum sample preparation:
[0029] (1) All serum samples were thawed in an ice bath for 30-60 minutes and then centrifuged for 5 minutes. Take 10ul of serum and 20ul of denaturation buffer U9 (9M Urea, 2% CHARPS, 50mM Tris-HCl PH9) and mix thoroughly. Medium shaking for 30 minutes;
[0030] (2) Carefully take out the SAX chip and mark it, put it into the biochip processor, add 200ul SAX binding buffer to each well and shake twice to pretreat the chip;
[0031] (3) Take 10ul of the sample and 110ul of SAX binding buffer (50mM Tris-HCl pH9) solution and mix quickly to avoid bubbles, add 100ul of the processed serum sample to each well, and shake at 4°C for 60 minutes;
[0032] (4) After serum samples, add 200ul SAX binding buffer to each well, shake twice at room temperature, then add 200ul deionized water to wash the chip twice;
[0033] (5) Take out the chip, add 0.5ul SPA to each well after micro-drying, and repeat once after micro-drying;
[0034] (6) On 5 machi...
Embodiment 2
[0049] 1. Serum sample information:
[0050] The 86 serum samples were all collected from the Second Affiliated Hospital of Zhejiang University School of Medicine from February 2003 to November 2003 with the informed consent of the hospital ethics committee and the patients. Non-gastric cancer healthy volunteers with gastroscopic examination and pathologically confirmed chronic superficial gastritis or chronic atrophic gastritis without a history of malignant tumors. Among the patients, there were 31 males and 14 females, with an average age of 56.5 years (36-76 years old). UICC tumor stages: 14 cases of stage I, 6 cases of stage II, 10 cases of stage III, and 15 cases of stage IV. The control group was paired by age and sex, including 28 males and 13 females, with an average age of 52.6 years (38-73 years). All serum samples were drawn before treatment on an empty stomach in the morning, and the serum was stored in a -80°C low-temperature refrigerator.
[0051] 2. Instrumen...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| laser intensity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com