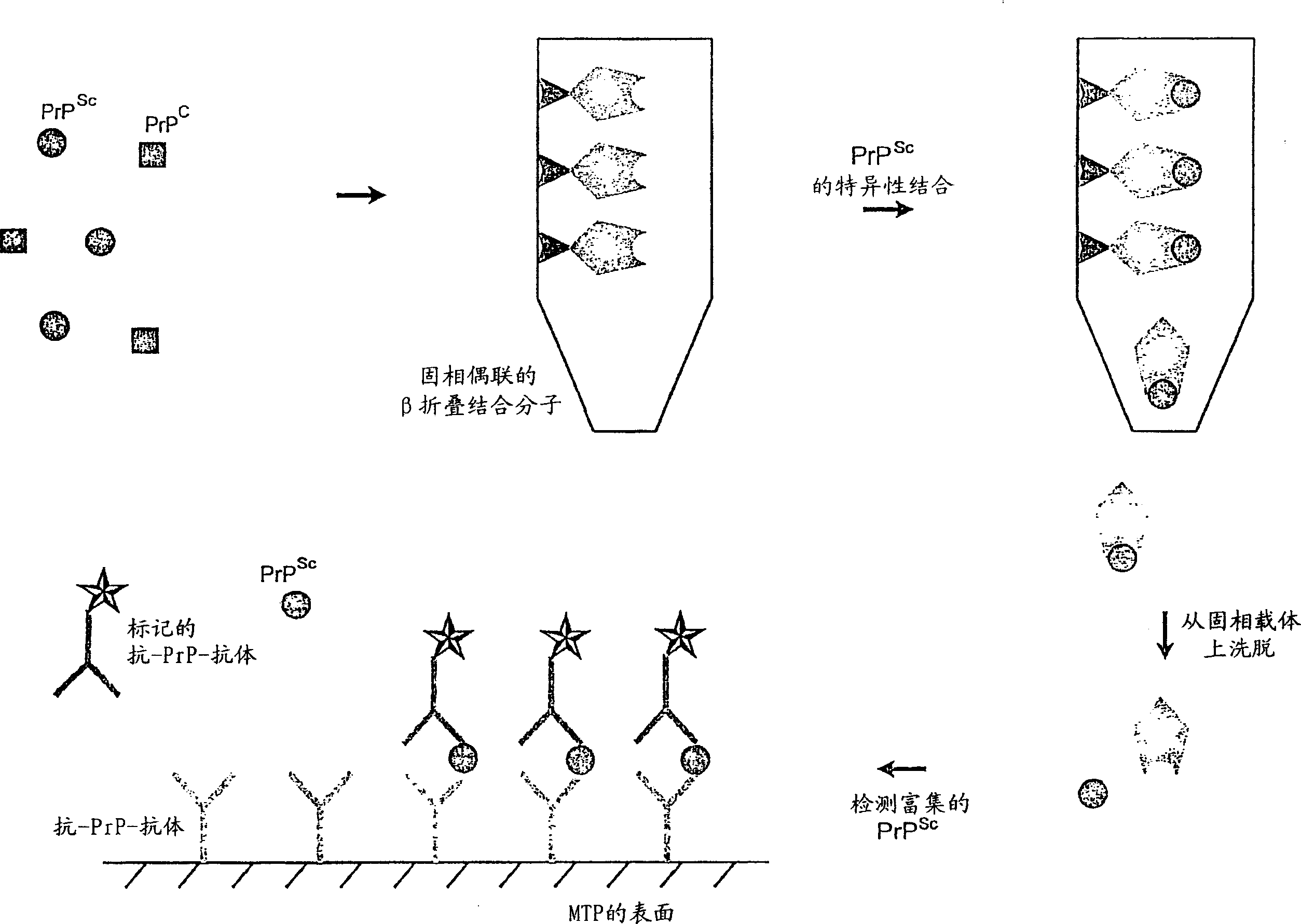
Method for enriching and tracking pathologic modified prions-proteins (PrP)
A prion, pathology technology, applied in chemical instruments and methods, biochemical equipment and methods, disease diagnosis, etc., can solve the problems of limited protein binding ability, high cost, incompatibility, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1: Isolation of PrP from brain homogenate using different peptides in the form of MTP Sc
[0045] Microtiter plate (MTP) (Nunc-Immuno TM Plate Maxisorp TM Surface, F96 (Nunc, Roskilde, Denmark)), was coated with the peptides listed in Table 1 (see figure 2 ). The coating was performed by co-incubating each well with 100 μl of peptide solution (10 μg / ml in 0.1 M carbonate buffer, pH 9.6) at 4° C. for 16 hours. The liquid was aspirated by vacuum and the wells were washed three times with 300 μl of wash buffer (PBS (10 mM phosphate buffer, 0.15 M NaCl, pH 7.2); 0.05% Tween 20)). Free binding sites were blocked by incubation with 0.5% casein in wash buffer for 1 hour at room temperature.
[0046] After a wash phase (300 μl wash buffer / well), the coated MTP was covered with foil and mixed with 100 μl PrP-containing Sc Samples / well (brain homogenates from BSE positive animals, which were prepared on Platelia OD > 4.0; a positive finding confirmed by immu...
Embodiment 2
[0050] Example 2: PrP Binding to KLVEF in the MTP Model Sc Elution of
[0051] PrP Sc The binding to the capture molecule is very strong and requires relatively strong conditions to elute PrP from the solid support Sc . Elution conditions were detected using the MTP model.
[0052] As in Example 1, MTP was coated with peptide 2 (KLVEF) and filled with PrP-containing Sc brain homogenate (Platelia Medium OD > 3.0). After washing 3 times with 300 μl of washing buffer (PBS; 0.05% Tween 20) per well, 100 μl of potent elution buffer (see Table 2) was added per well and incubated at room temperature for 5 minutes. Subsequently, the liquid is removed and the Platelia BSE Detection Kit (Bio-RadLaboratories, Hercules, USA) measures the PrP eluted in the elution buffer Sc The amount of and PrP left on the MTP Sc amount. Comparing the elution efficiencies (Table 2) shows that only the wash-containing buffer (containing 5% SDS) is suitable for the complete elution of PrP...
Embodiment 3
[0056] Example 3: Covalent binding of peptide KLVFF to EAH Sepharose
[0057] If bound via the amino group, the peptide will be fixed at the N-terminus and side chain (Lys), which disrupts the three-dimensional structure. To this end, the specific binding of the β-sheet binding molecule KLVFF to the solid support can be achieved through the carboxyl group at the C-terminus of the peptide. EAH-Sepharose 4B (Amersham Pharmacia Biotech, Uppsala, Sweden) was used as carrier material.
[0058] To prepare for the binding reaction, the EAH-Sepharose gel was washed with 0.5M NaCl and any excess liquid was completely removed. The ligand was a pentapeptide with the sequence KLVEF, which was dissolved in water at a final concentration of 5 mg / ml and the pH was adjusted to 4.5 with HCl. Resuspend the gel in ligand solution (1 part gel + 2 parts ligand solution) and add EDC (N-ethyl-N'-(3-dimethylaminopropyl)-carbodiimide ) to make the final concentration 0.1M. The binding reaction w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com