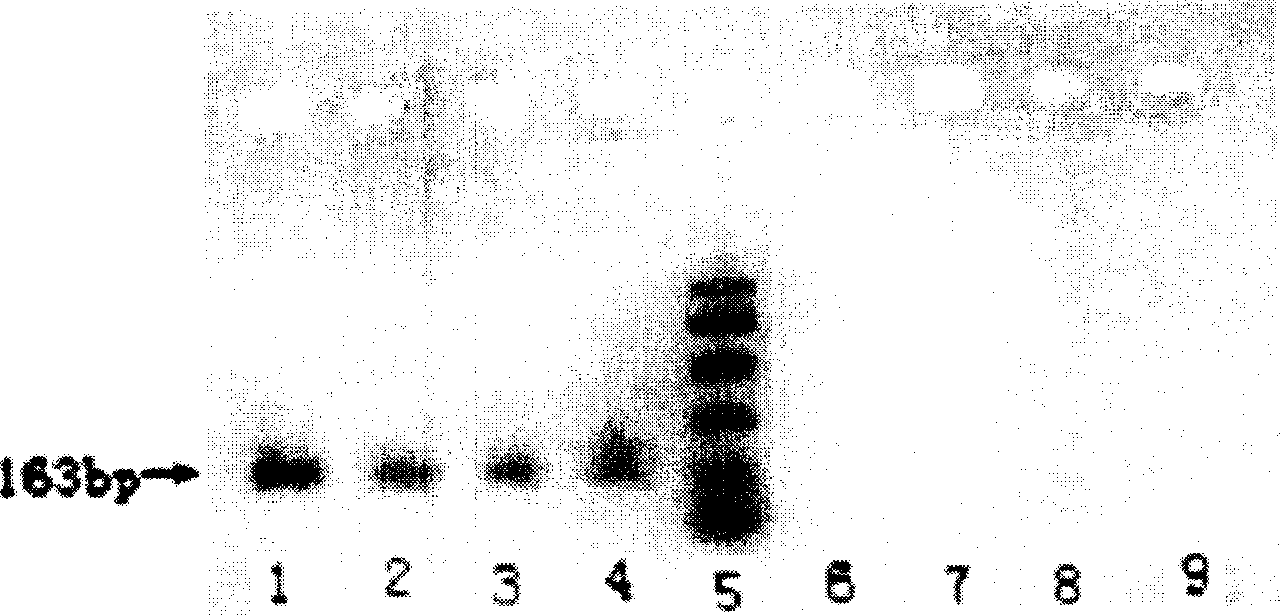PCR amplification primer, kit and use thereof in identification of mammal sex
A technology for amplification primers and kits, which is applied in the field of PCR amplification primers, kits, and gender identification of mammals, can solve the problems of difficult implementation, low identification accuracy, and long time required, etc. To achieve the effect of avoiding pollution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
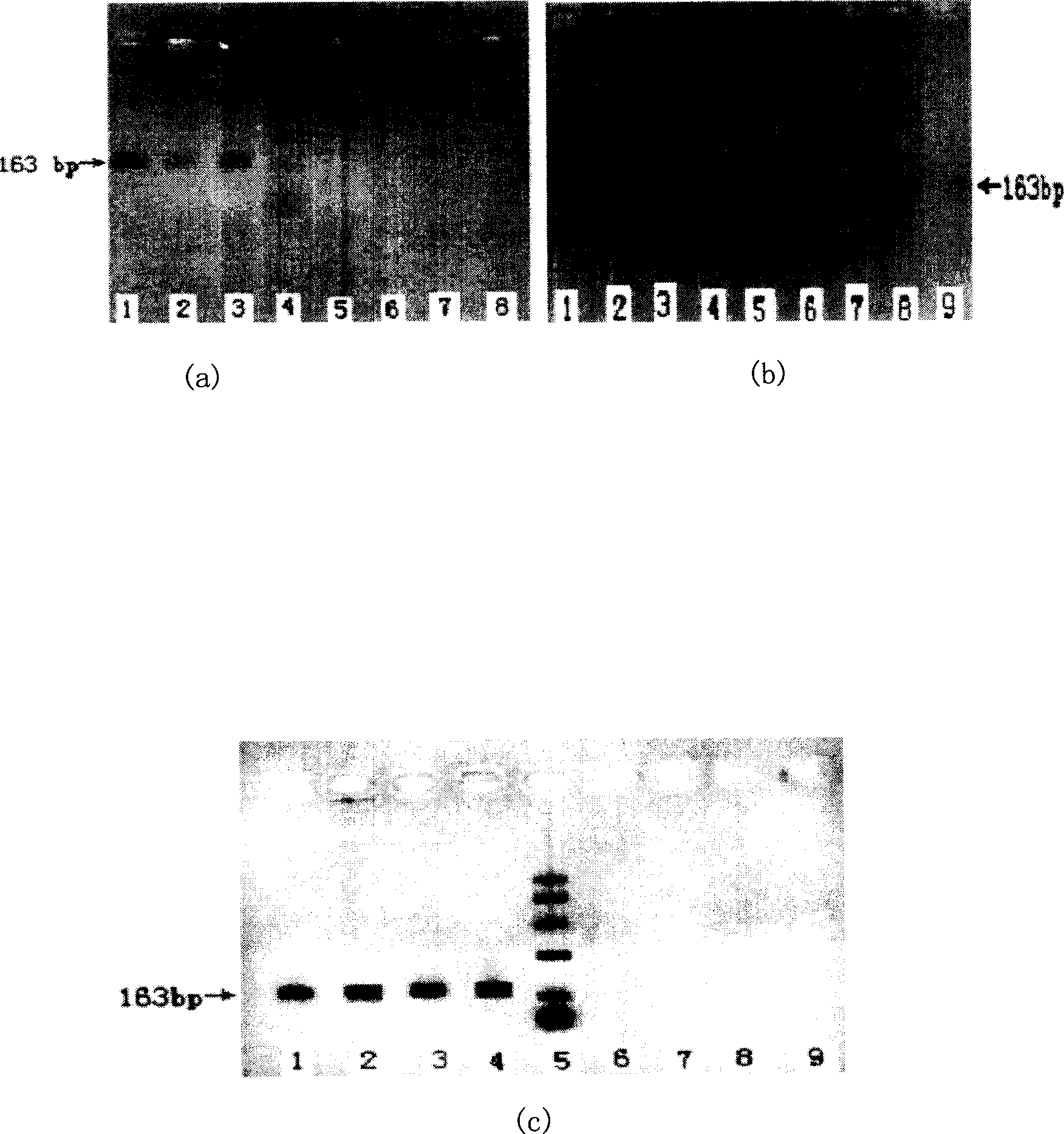
[0065] Gender detection using animal genome DNA
[0066] Extraction, purification and quality inspection of genomic DNA from animal tissues
[0067] 1. Cut 0.5 mg of fresh animal muscle tissue or liver tissue into pieces and place in 4.5 mL of DNA extraction buffer (10 mmol / LTris-HCl, 0.1mol / LEDTA, 20 μg / mL RNase, 0.5% SDS pH8.0) In the homogenization tube, the homogenization tube was placed in a beaker with ice cubes, and an electric glass homogenizer was used to homogenize at a medium speed for about 90 sec. All the homogenate was poured into two 5mL centrifuge tubes, and the centrifuge tubes were placed in a beaker with ice cubes, and crushed with an ultrasonic cell pulverizer (power 400W, ultrasonic time 3 sec, interval time 4 sec, working frequency 15 times). EDTA refers to ethylenediaminetetraacetic acid.
[0068] 2. After crushing the cells, quickly place the centrifuge tube in a constant temperature shaker in a water bath and incubate at 37°C for 1 h.
[0069] 3. Ad...
example 2
[0088] Extraction, purification and quality detection of genomic DNA from animal serum
[0089] 1. Collect about 6mL of fresh blood and add it to a centrifuge tube filled with 1mL acid citrate glucose solution B (ACD solution—0.48g citric acid, 1.32g sodium citrate, 1.47g glucose, add water to 100mL). Blood can be stored at 0°C for several days or at -70°C for long-term storage before DNA preparation.
[0090] a. Fresh blood (2mL) was centrifuged at 4500r / min for 15min, the upper layer of plasma was discarded, and the light yellow layer was carefully transferred to a new centrifuge tube with a pipette, and centrifuged again at 4500r / min for 15min. After centrifugation, resuspend the buffy layer in 1.5 mL of DNA extraction buffer, incubate at 37° C. for 1 h, and follow steps 3-11 in Example 1.
[0091] b. Refrigerated blood (2mL), thawed at room temperature, transferred to a new centrifuge tube, washed with an equal volume of phosphate-buffered saline PBS (8g NaCl, 0.2g KCl, 1...
example 3
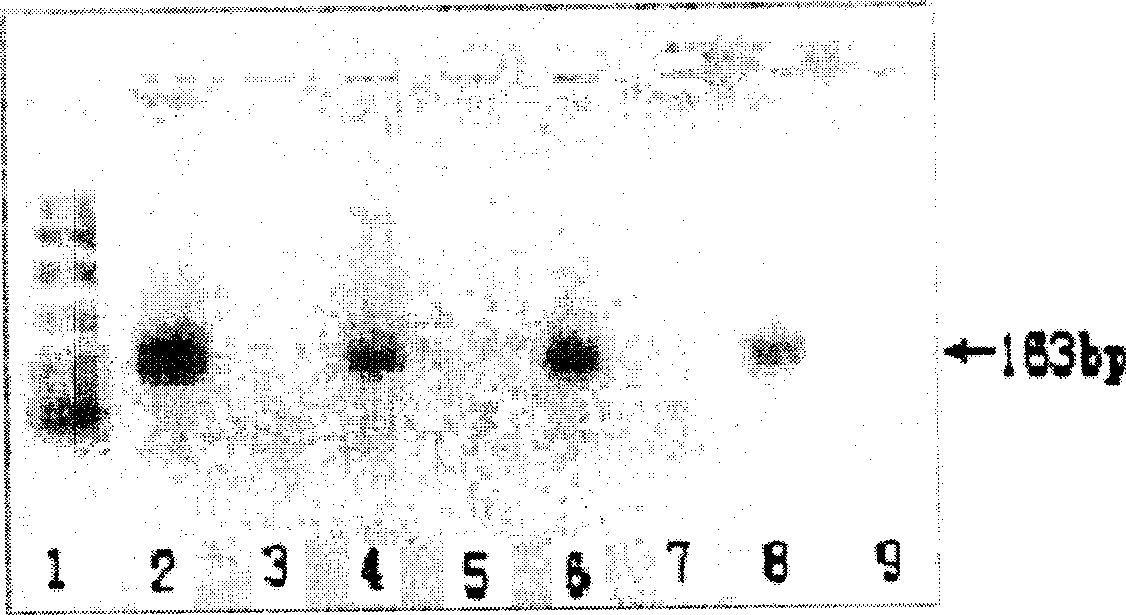
[0098] Methods of sex determination using embryos
[0099] 1. Evaluation and size determination of embryos
[0100] Embryos detected under a solid microscope were washed with PBS for 3 times and placed in a sterile room for 0.5 hours. In the culture medium, the developmental stage, embryo age and grade were evaluated.
[0101] 2. Treatment of Embryos
[0102] Collect 1~2 cells of mammalian embryos, first rinse with 0.145mol / L NaCl twice, then add embryo treatment solution (10mmol / LTris-HCl (pH8.3), 50mmol / L KCl, 2mmol / L MgCl 2 , 0.45% NP-40, 0.45% Tween-20, 0.1μg / μL proteinase K), so that the total volume is 20μL, the embryo and its treatment solution are reacted in a 55°C water bath for 60min, and then the embryo lysate is directly used as a PCR reaction template.
[0103] 3. Amplification reaction
[0104] According to the number of samples to be amplified n (n = the number of processed embryo samples + 2), take PCR reaction solution n×29.μL and Taq DNA polymerase n×0.4 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com