Pseudo monads pseudoalcaligenes gene promoter
A Pseudomonas, promoter technology, applied in genetic engineering, plant genetic improvement, biochemical equipment and methods, etc., can solve problems such as low transcription efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Pseudomonas pseudoalcaligenes total DNA extraction
[0048] Pseudomonas pseudoalcaligenes was activated overnight in 2ml LB, inoculated in 50ml broth medium at a ratio of 1:100, and cultured for 12hr. Put the bacteria into a 50ml centrifuge tube, centrifuge at 3500rmp for 10min, remove the supernatant, turn the centrifuge tube upside down on a paper towel and let the supernatant flow out at night; add a small amount of lysate to suspend the bacteria, and then add lysate to a total volume of 4ml , add 1ml SDS, mix well, 60°C for 10min; add equal volumes of phenol, phenol / chloroform extract, and chloroform to extract once; take the supernatant, add 1 / 10 volume of 3M potassium acetate (pH4.8), mix Mix well, then add 2 times the volume of 100% ice-cold ethanol along the tube wall, shake gently until a filamentous precipitate appears; centrifuge at 2000rpm for 5 minutes, remove the supernatant; add 1ml of 70% ethanol, wash the precipitate once, and let it dry naturally at ro...
Embodiment 2
[0050] produce DNA fragments
[0051] Use Pseudomonas pseudoalcaligenes total DNA extract (1 μ g) to pass through in (100 μ l volume) 100 mM NaCl, 50 mM Tris-HCl (pH7.5), 10 mM MgCl 2 , Dithiothreitol in 1mM was digested with Sau3A I [Bao Biological Engineering (Dalian) Co., Ltd. (TAKARA)] to generate a DNA fragment of Pseudomonas-like alcaligenes. The mixture was incubated at 37°C for 120 minutes and quenched with 10 mM EDTA. The Sau3A1 fragment was precipitated from the digestion mixture with ethanol and washed with ethanol.
Embodiment 3
[0053] Transformation of E. coli and selection of transformants
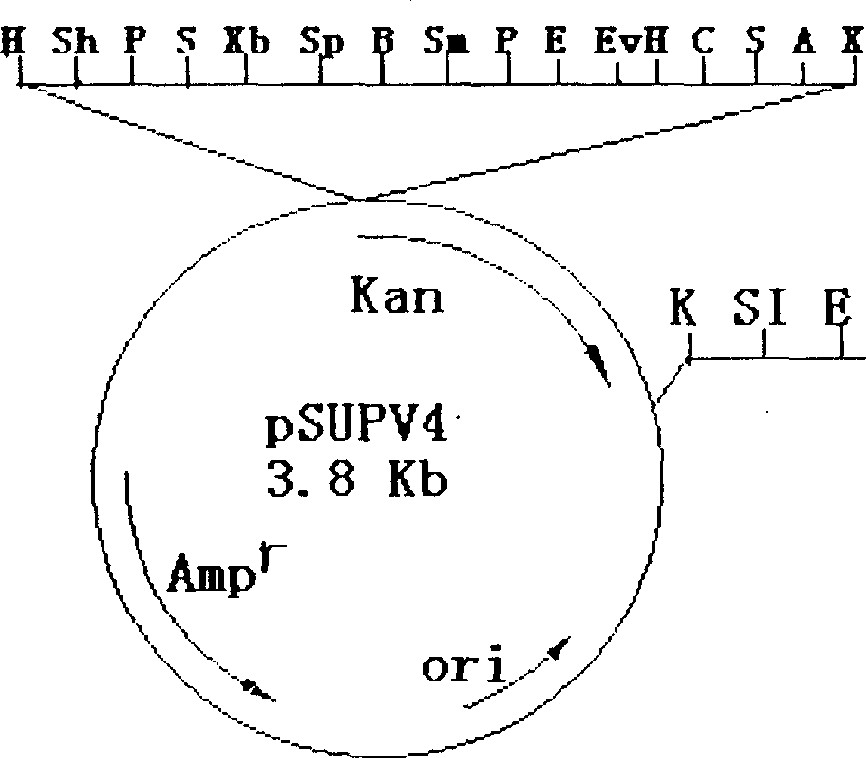
[0054] The Sau3AI viral fragment produced as described in Example 1 was cloned into plasmid pSUPV4 by the shotgun cloning method described in Sambrook et al., 1989, in Cloning. Plasmid promoter probe vector pSUPV4 (AP r ) was constructed by the Laboratory of Molecular Biology, School of Life, Sichuan University (Zhang Yizheng et al. Journal of Sichuan University (Natural Science Edition), 1998, 35(2): 263-267.). The map of pSUPV4 is shown in Figure 1 . DNA preparation, fill-in reactions and ligation were performed as described by Sambrook et al. (supra) or according to the manufacturer's instructions.
[0055] The DNA fragment of Pseudomonas pseudoalcaligenes was ligated with 1 μg of pSUPV4 pre-treated with BamHI and calf intestinal alkaline phosphatase (CIP) to generate recombinant plasmids PA1; PA2; PA3; PA4: PA5; PA6; PA7; PA8; PA9 . These 9 are related to the heterologous kanamycin resistance marker gene...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com