Fusion protein, double-deaminase-mediated base editing system containing same and application of double-deaminase-mediated base editing system
A fusion protein and base editing technology, applied in the field of base editing systems mediated by double deaminase, can solve the problems of low editing efficiency, unsatisfactory, and low fusion protein base editing efficiency, and achieve high editing efficiency. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
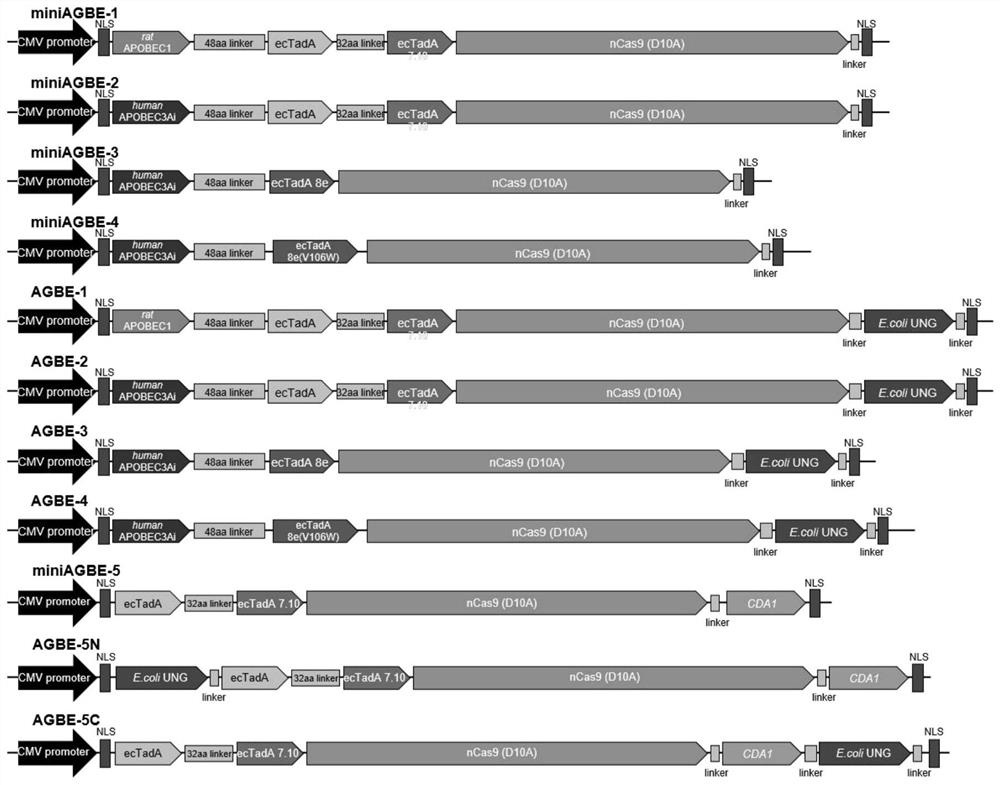
[0114] This example provides fusion proteins AGBE-1, AGBE-2, AGBE-3, AGBE-4, miniAGBE-1, miniAGBE-2, miniAGBE-3, miniAGBE-4, miniAGBE-5, AGBE-5C and AGBE-5N.
[0115] The preparation method of the fusion protein AGBE-1 comprises: using ABEmax (pCMV-bis-bpNLS-wild typeecTadA-ecTadA*7.10-nCas9(D10A)-bis-bpNLS-poly(A)) as the basic carrier, through a 48 The connecting peptide sequence of amino acid length connects the APOBEC1 (R33A) sequence of mouse origin to the N-terminal of ABEmax, and then connects the uracil DNA glycosylase (E.coli UNG) sequence is connected between the nuclease (nCas9 (D10A)) sequence and the nuclear localization signal bis-bpNLS (NLS) sequence to construct the fusion protein AGBE-1 expression vector, the structure diagram is shown in figure 1 , the sequence of the fusion protein AGBE-1 is SEQ ID NO.10.
[0116] The preparation method of the fusion protein AGBE-2 comprises: on the basis of the AGBE-1 expression vector, replacing the cytosine deaminase AP...
Embodiment 2
[0123] In this example, AGBE-1, AGBE-2, AGBE-3, AGBE-4, miniAGBE-1, miniAGBE-2, miniAGBE-3 and miniAGBE-4 were used to perform base editing in HEK293 cells.
[0124] In the experimental group, AGBE-1, AGBE-2, AGBE-3, AGBE-4, miniAGBE-1, miniAGBE-2, miniAGBE-3 and miniAGBE-4, and sgRNA at specific sites were used in a mass ratio of 3:1 (6μg: 2μg) co-transfected into HEK293 cell line (0.8×10 6cell amount / group). In the negative control group, only sgRNA at a specific site was added, and the amount of a single carrier was the same as that in the experimental group. In the blank control group (WT) group, only the corresponding electroporation resuspension solution R was added. After 72 hours of transfection, the cells were harvested for lysis and the genome was extracted. Using the cell lysate as a template, the target fragment containing the target site was amplified by PCR reaction, and the overall base editing status of the site was identified by Sanger sequencing.
[0125] ...
Embodiment 3
[0129] This application uses miniAGBE-5, AGBE-5N and AGBE-5C for base editing in HEK293 cells.
[0130] In the experimental group, miniAGBE-5, AGBE-5N and AGBE-5C, as well as specific site sgRNA were co-transfected into HEK293 cell line (0.8×10 6 cell amount / group), only sgRNA at a specific site was added to the negative control group, and the dosage of a single carrier was consistent with that of the experimental group. Only the corresponding electroporation resuspension solution R was added to the blank control group (WT) group, and 72 hours after transfection , collect the cells for lysis, extract the genome, use the cell lysate as a template, use PCR reaction to amplify the target fragment containing the target site, and identify the overall base editing status of the site by Sanger sequencing.
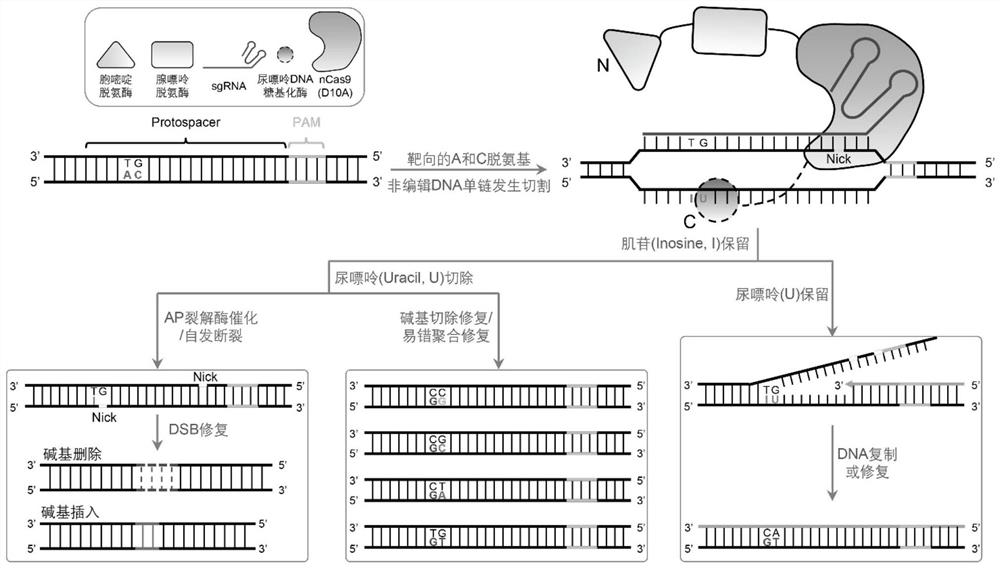
[0131] The frequency of A and C base transitions at endogenous loci in HEK293 cells is as follows Figure 6 As shown, the abscissa indicates the specific position of adenine A an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com