Engineered trans-enoyl COA reductases and methods of making and using same
A trans-enoyl, engineering technology, applied in the field of engineered trans-enoyl COA reductase and its preparation and use, can solve the problems of increasing the cost and complexity of biosynthetic compounds, reducing product efficiency or yield, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
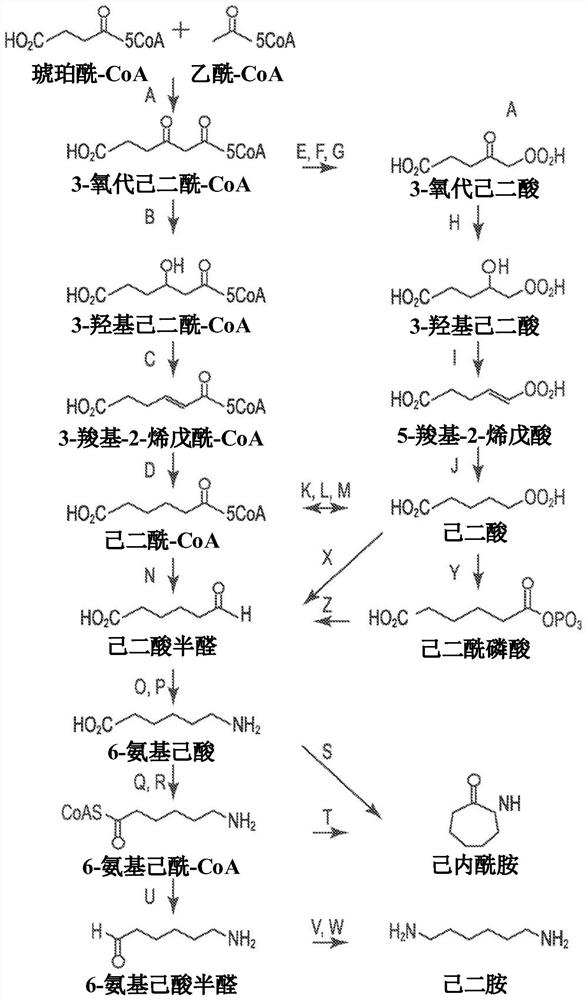
[0194] Example 1. Identification of 5-carboxy-2-pentenoyl-CoA reductase.
[0195] Genes encoding 5-carboxy-2-pentenoyl-CoA reductase (Ter) were identified from the literature and bioinformatically from public databases using the Basic Local Alignment Search Tool (BLAST) (Table 3). Genes encoding each reductase were synthesized, expressed in E. coli, and 5-carboxy-2-pentenoyl-CoA (CPCoA) reductase activity was assessed using an enzyme coupled assay.
[0196] The genes encoding the TER enzyme candidates of Table 3 were cloned into low copy vectors under constitutive promoters and the constructs were transformed into E. coli using standard techniques. The transformants were cultured overnight at 35° C. in LB medium containing antibiotics, and then the cells were centrifuged at room temperature at a speed of 15000 rpm. To prepare lysates, remove supernatant and resuspend Ter gene-expressing E. coli cells in chemical lysis solution containing lysozyme, nuclease, and 10 mM DTT. Us...
Embodiment 2
[0207] Example 2. In vivo activity of 5-carboxy-2-pentenoyl-CoA reductase.
[0208] Two identified 5-carboxy-2-pentenoyl-CoA reductases were also tested in an in vivo assay in which they were modified to express the enzyme encoding 3-oxoadipyl-CoA thiolase (Thl), 3 Escherichia coli strains containing the genes for -oxoadipyl-CoA dehydrogenase (Hbd) and 3-oxoadipyl-CoA dehydratase ("crotonase" or Crt) were used to contain 5-carboxy-2-pentane Construct transformation of the enoyl-CoA reductase gene. The gene encoding the TER enzyme of Candida tropicalis (SEQ ID NO:1, referred to as candidate #1) and the gene encoding the TER of Drosophila melanogaster (SEQ ID NO:24, candidate #24) were respectively cloned into low copy in the plasmid vector under a constitutive promoter. The amino acid sequences of Candida and Drosophila TER are about 40% identical; image 3 shown. The vector expressing the Ter gene was transformed into a Thl / Hbd / Crt E. coli strain. The transformants were t...
Embodiment 3
[0214] Example 3. Targeted mutagenesis of Ter gene.
[0215] Using structure-guided design, and then by codons at Q52, V105, S148, V149, T153, V301, T302 and N307, and Q11, A39, S48, S59, G97, S103, H104, N106, F107, I147, L152 , L156, R200, D201, R202, E303, K306, N308, and L316 codons to mutate the gene encoding the Candida TER enzyme (SEQ ID NO: 1) to generate the Candida TER enzyme encoding (SEQ ID NO: 1) gene variants. Similarly, structure-guided design was used followed by mutating the gene encoding the Candida TER enzyme (SEQ ID NO: 1) at the codons for S96, L98, V129, M274, T275 to generate Variants of the gene for the enzyme (SEQ ID NO: 1). Amino acid position substitutions were performed using degenerate primer sequences and PCR, wherein the altered gene sequence mixture was transformed into E. coli.
[0216] Transformants were tested in lysate assays as described in Example 1 and re-tested in lysate assays to provide assays that exhibited higher activity than wil...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com