Primer group, kit and detection method for detecting polymorphism of risk genes of blood hypercoagulation or venous thrombosis and application
A gene polymorphism and venous thrombosis technology, applied in the field of molecular biology, can solve the problems of poor primer specificity, unfavorable clinical promotion, and low sensitivity, and achieve the effect of complete evaluation system, wide coverage of genes, and accurate detection results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
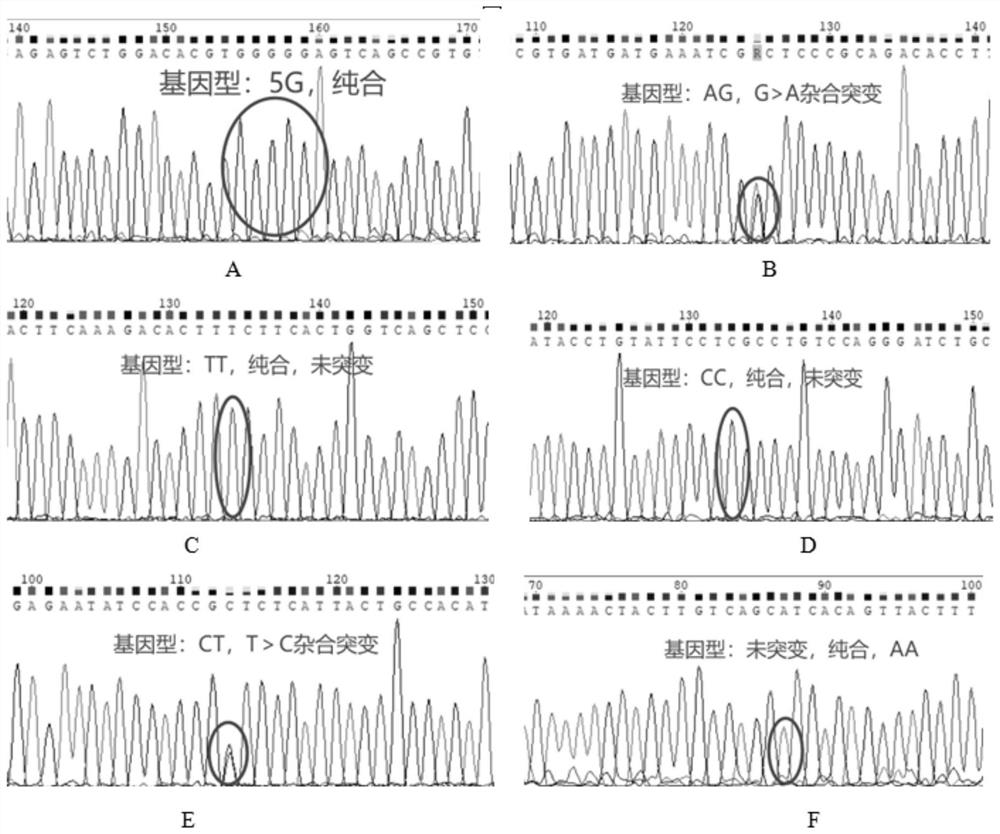
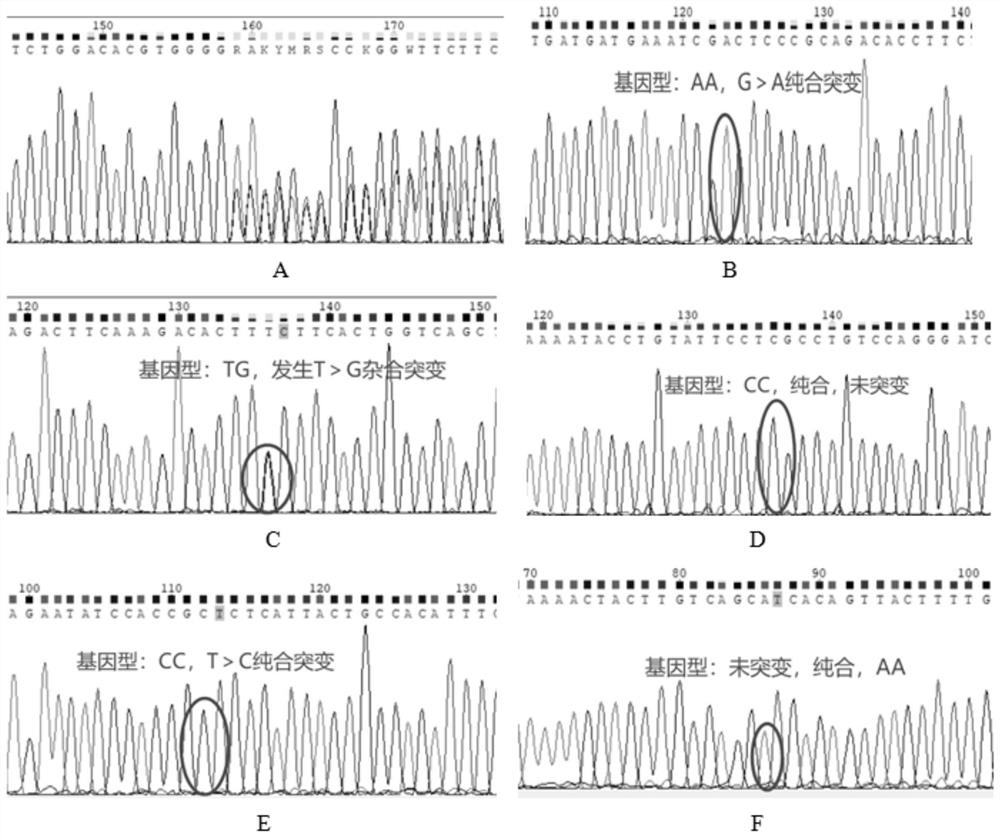
[0059] According to the polymorphic sites of PAI-1, MTHFR, F5, and TDAG8 (also called GPR65) genes in the dbSNP database, the dbSNP request number information, and the nucleotide sequences of each gene, design specific amplification primers.
[0060] Among them, the site PAI-1 (4G / 5G), the dbSNP access number is rs1799762; the site MTHFR (677C>T), the dbSNP access number is rs1801133; the site MTHFR (1298A>C), the dbSNP access number is rs1801131; For point F5 (41721G>A), the dbSNP accession number is rs6025; for the locus LINC01146(C>T)HIT1, the dbSNP accession number is rs1887289; for the locus GPR65(T>G)HIT2, the dbSNP accession number is rs3742704.
[0061] The partial amplified nucleotide sequence of each gene is as follows:
[0062] PAI-1 (4G / 5G): SEQ.ID.NO.1
[0063]
[0064] MTHFR(677C>T): SEQ.ID.NO.2
[0065]
[0066] MTHFR(1298A>C): SEQ.ID.NO.3
[0067]
[0068]
[0069] F5(41721G>A): SEQ.ID.NO.4
[0070]
[0071] LINC01146 (C>T) HIT1: SEQ.ID.NO.5 ...
Embodiment 2
[0080] According to embodiment 1, a primer set for detecting blood hypercoagulability or venous thrombosis risk gene polymorphism, the primers include:
[0081] (1) Primers for detecting PAI-1 (4G / 5G) polymorphisms, including primers shown in SEQ.ID.NO.7 and SEQ.ID.NO.8;
[0082] (2) primers for detecting MTHFR (677C>T) polymorphism, including primers shown in SEQ.ID.NO.9 and SEQ.ID.NO.10;
[0083] (3) primers for detecting MTHFR (1298A>C) polymorphism, including primers shown in SEQ.ID.NO.11 and SEQ.ID.NO.12;
[0084] (4) Primers for detecting F5 (41721G>A) polymorphism, including primers shown in SEQ.ID.NO.13 and SEQ.ID.NO.14;
[0085] (5) Primers for detecting LINC01146(C>T)HIT1 polymorphism, including primers shown in SEQ.ID.NO.15 and SEQ.ID.NO.16;
[0086] (6) Primers for detecting GPR65(T>G)HIT2 polymorphism, including primers shown in SEQ.ID.NO.17 and SEQ.ID.NO.18.
[0087] In this embodiment, a detection kit is prepared for detecting the polymorphism of blood hyperc...
Embodiment 3
[0102] In this example, using the genome fragments extracted from the blood cells of healthy pregnant women as templates, 6 pairs of primers designed for SNP sites related to venous blood hypercoagulability were amplified, and on this basis, a method for detecting human venous thrombosis susceptibility based on Sanger sequencing was established. . It mainly includes the following steps and reagents:
[0103] In this example, the primers and kits described in Examples 1 and 2 were used for detection.
[0104] In the kit, 2×Taq Master Mix as an amplification reagent contains Taq DNA Polymerase, dNTP and an optimized buffer system, which can be amplified only by adding primers and templates. The protective agent added to the amplification system makes the 2×Master Mix maintain stable activity after repeated freezing and thawing.
[0105] (1) Genomic DNA extraction: Use Kangwei CW2087 Blood Genomic DNA Column Small Extraction Kit to extract 200 μL EDTA anticoagulated blood. The ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com