Method for synthesizing propionic acid by using threonine and recombinant bacterium used in method
A technology of recombinant bacteria and threonine, applied in the field of recombinant bacteria, can solve problems such as global overcapacity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
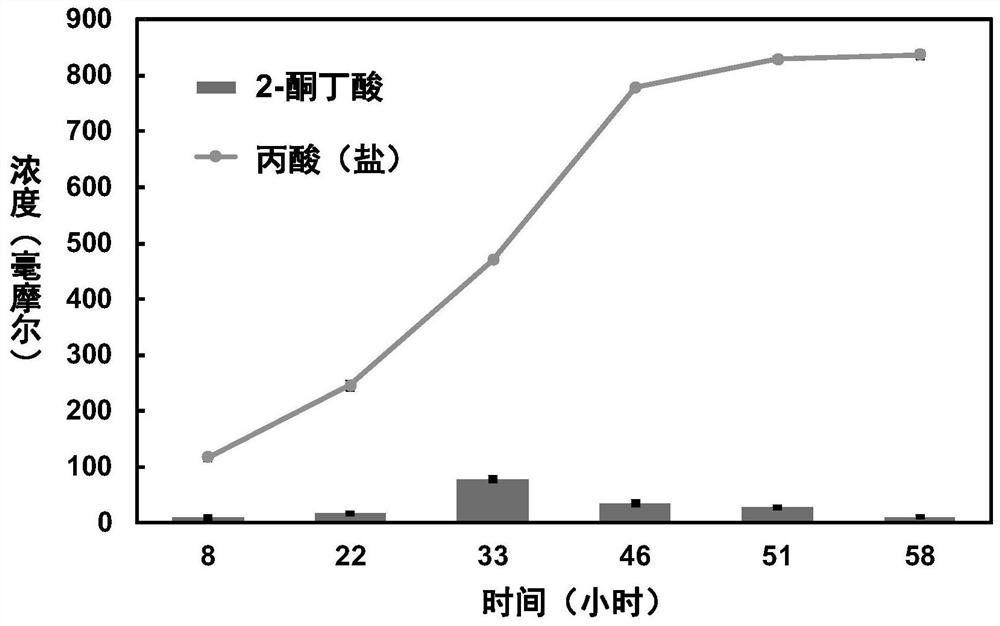
Examples
Embodiment 1
[0128] Embodiment 1, the preparation of recombinant bacteria PS30
[0129] 1.1 Knockout of threonine aldolase ltaE
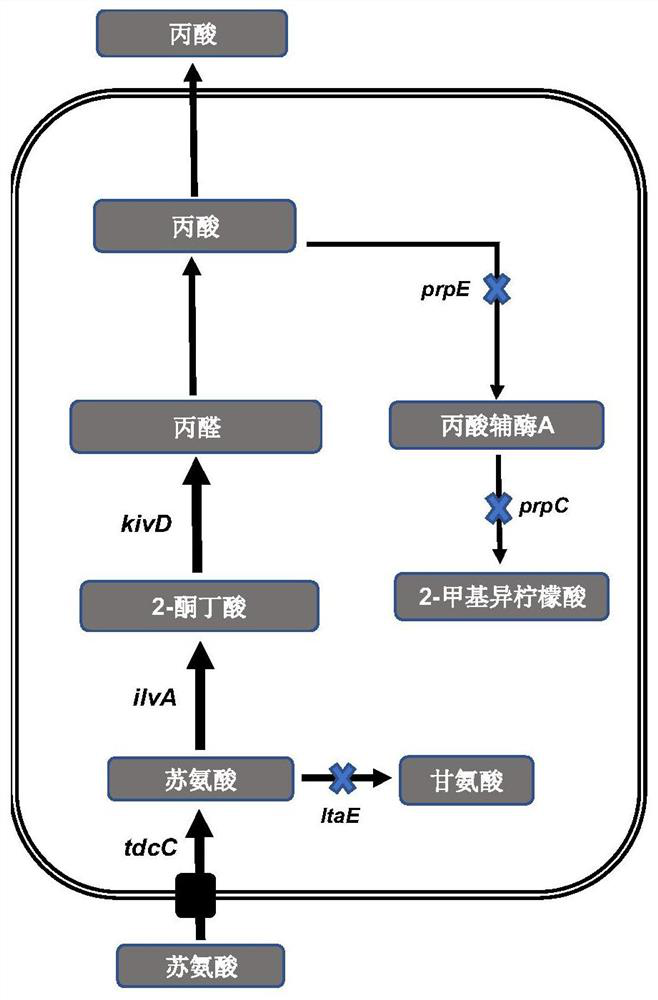
[0130] Starting from Pseudomonas putida KT2440 (ATCC 47054), the threonine aldolase gene ltaE was knocked out by homologous recombination (Gene ID: 1044018, discontinued on 2-Apr-2020; Genome: NC_002947.4:c386535 -385495), which blocks the threonine to glycine pathway pathway. The amino acid sequence of threonine aldolase is shown in WP_010951681.1 (2017-5-14) in NCBI. The specific steps are as follows:
[0131] 1.1.1. Construction of knockout plasmid pK18-ltaE
[0132] Using the pK18 plasmid (ATCC 87097) as a template, it was cut with two restriction enzymes, BamHI and HindIII, to obtain a 6022bp DNA linearized fragment.
[0133] Select 500bp upstream and downstream of the ltaE gene (SEQ ID No.2 1-500 and SEQ ID No.2 2163-2662), and design primers ltaE-up-F / ltaE-up-R and ltaE-down-F / ltaE-down-R, using the genomic DNA of Pseudomonas putida KT2440 as a temp...
Embodiment 2
[0162] Embodiment 2, the preparation of recombinant bacterium PS31
[0163] 2.1. Genome overexpression of threonine deaminase gene ilvA derived from Escherichia coli
[0164] The amino acid sequence of threonine deaminase derived from Escherichia coli is shown in EEV5737228.1 (20200406) in NCBI. The preparation steps are as follows:
[0165] 2.1.1. Construction of overexpression ilvA plasmid
[0166] Extract genomic DNA from Escherichia coli (ATCC 700926), amplify ilvA gene with primer ilvA-F / ilvA-R, obtain the DNA fragment of 1545bp (the nucleotide sequence of coding chain is as SEQ ID No.2 the 617th-2161st position shown). Plasmid pUCP18 (BioVector, Cloning vector pUCP18) was cut with restriction endonucleases BamHI and HindIII to obtain a 5465bp DNA linearized fragment. The 1545bp DNA fragment ilvA and the 5465bp pUCP18 plasmid DNA linear fragment were ligated together by the Gibson method, and transformed into Escherichia coli DH5α (transgen biotech, CD201-01), identif...
Embodiment 3
[0188] Embodiment 3, the preparation of recombinant bacterium PS32
[0189] 3.1. Overexpression of 2-ketoisovalerate deacidase derived from Lactococcus lactis
[0190] The amino acid sequence of 2-ketoisovalerate decarboxylase (kivD) derived from Lactococcus lactis is as WP_012897921.1 (20190728) in NCBI, and the preparation method is as follows:
[0191] Genomic DNA was extracted from Lactococcus lactis (NCDO 2118), and the kivD gene was amplified with primers kivD-F / kivD-R to obtain a 1647bp DNA fragment (the coding sequence of the coding strand is shown in SEQ ID No.3). Plasmid pUCP18 (BioVector, Cloning vector pUCP18) was cut with restriction endonucleases BamHI and HindIII to obtain a 5465bp DNA linearized fragment. The 1647bp DNA fragment kivD and the 5465bp pUCP18 plasmid DNA linear fragment were ligated together by the Gibson method, and transformed into Escherichia coli DH5α (transgen biotech, CD201-01), identified with primers pucp18-F / pucp18-R, and selected the tar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com