Three-dimensional DNA walker and application thereof in tumor exosome detection
A tumor exosome and walker technology, applied in the field of biochemistry, can solve the problems of complex orbital fabrication, low synthesis yield, and high synthesis cost, and achieve the effect of simplifying the experimental operation process, low cost, and strong anti-false positive effect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
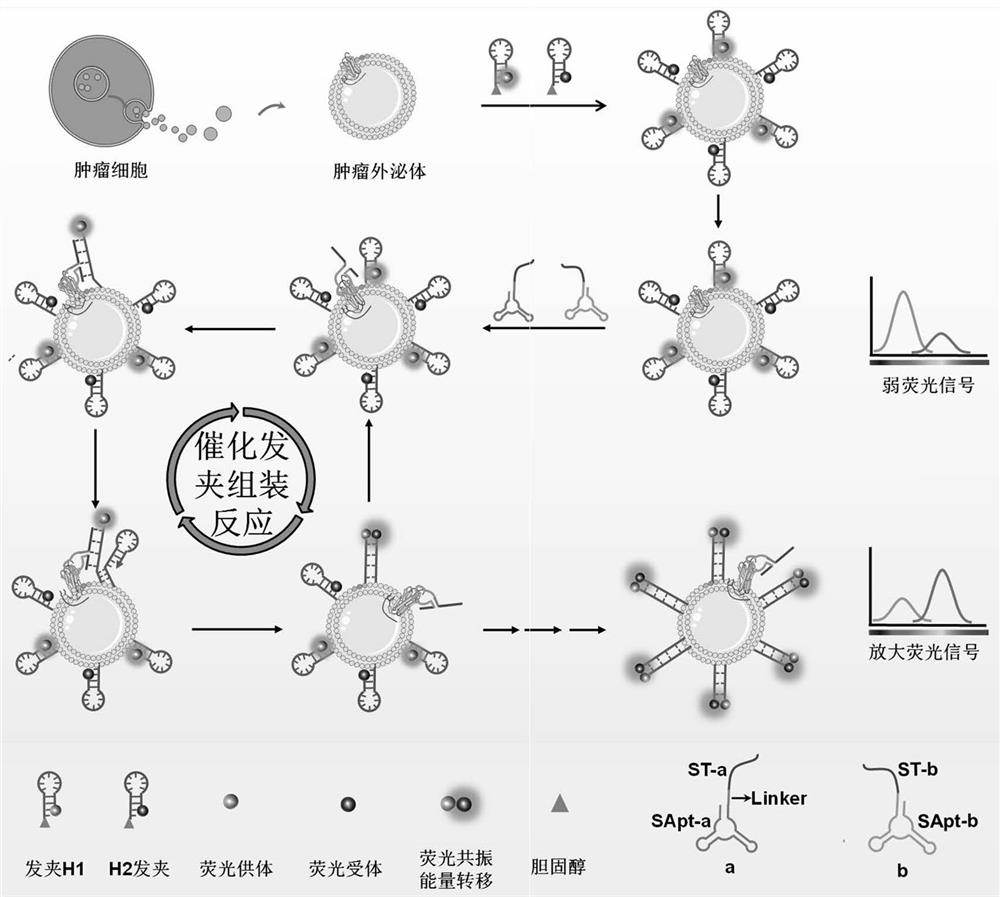
[0038] A recognition probe for exosomes of SMMC-7721 liver cancer according to the present invention includes split probe a, split probe b, hairpin H1 and hairpin H2.
[0039] The cleavage-type probe a includes a cleavage-type Aptamer sequence Apt-a, a cleavage-type trigger sequence Ta and a linker fragment linker. Among them, Apt-a and Ta are connected to split-type probe a through linker.
[0040] Split-type probe b includes Apt-b and Tb. Apt-b connects with Tb to form split probe b.
[0041] The split-type nucleic acid aptamers Apt-a and Apt-b in this example are formed by splitting a complete Aptamer sequence into two parts, which can be specifically recognized and assembled on the target protein on the surface of exosomes to form the same complete Aptamer has a similar recognition structure, a DNA fragment with specific recognition ability for SMMC-7721 liver cancer exosomes.
[0042] Wherein the nucleotide sequence of Apt-a is shown in SEQ ID NO.1:
[0043] 5'-acggac...
Embodiment 2
[0062] An application of the liver cancer exosome recognition probe of Example 1 in the detection of SMMC-7721 liver cancer exosomes.
[0063] Its application method is:
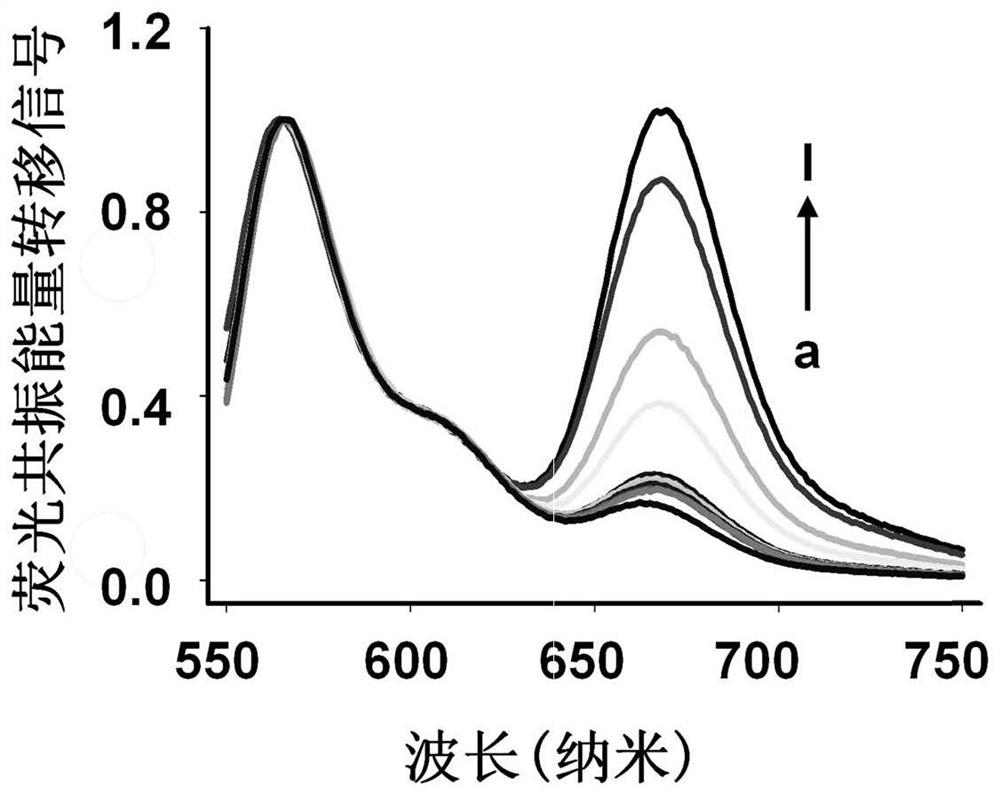
[0064] Add different concentrations of SMMC-7721 exosomes to Tris-HCl containing cholesterol-modified Cy3-H1 (200nM) and Cy5-H2 (200nM), and incubate at 37°C for 0.5h to ensure that the catalytic hairpin probe passes through cholesterol The interaction with the phospholipid bilayer sufficiently anchors the exosome membrane to prepare a three-dimensional track that catalyzes hairpin assembly. Then add 50nM cleave-type probes (50nM cleave-type probe a and 50nM cleave-type probe b) to the above reaction system, react at 37°C for 2.5h, and use steady-state near-infrared fluorescence after the fluorescence is stable. A spectroscopic system (QM40-NIR) records the fluorescence spectrum, the excitation wavelength is 530nm, the emission wavelength is 550-750nm, and the slit widths for both excitation and emission ar...
Embodiment 3
[0067] Investigate the detection specificity of the recognition probe of liver cancer exosomes in Example 1 to SMMC-7721 liver cancer exosomes:
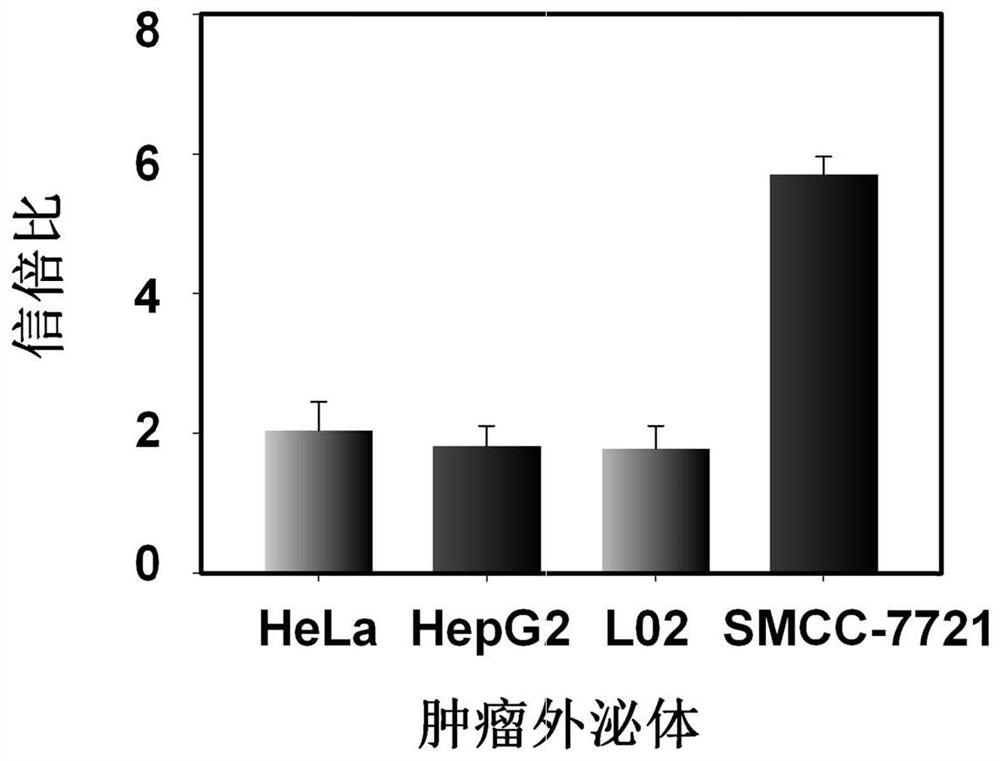
[0068] Add liver cancer SMMC-7721 exosomes to 100 μL Tris-HCl buffer solution containing Cy3-H1 (200 nM) and Cy5-H2 (200 nM), and incubate at 37 ° C for 0.5 h, until the hairpin probe is fully anchored in the exosomes After the phospholipid bilayer was coated, 50nM split probe was added to the reaction system and reacted at 37°C for 2.5h. After the fluorescence was stable, the fluorescence spectrum was recorded with a steady-state near-infrared fluorescence spectroscopy system (QM40-NIR). In addition, the buffer solution containing control exosomes such as human liver cancer HepG2 exosomes, human cervical cancer HeLa exosomes, and human normal liver cell L02 exosomes were tested, and the operation was as described above. For test results, see image 3 .
[0069] From image 3 It can be seen that the fluorescence resonance energy t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com