PCR primer group, kit and application thereof
A primer set and primer pair technology, applied in the field of medical diagnosis, can solve the problems of undetectable, difficult to detect, expensive and other problems of SNP chips
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
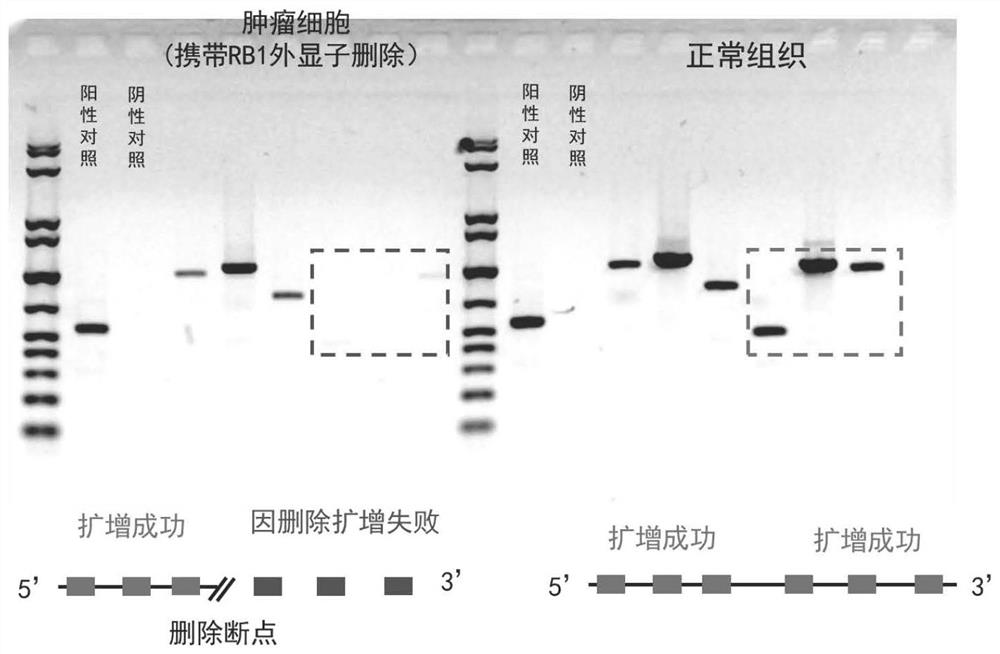
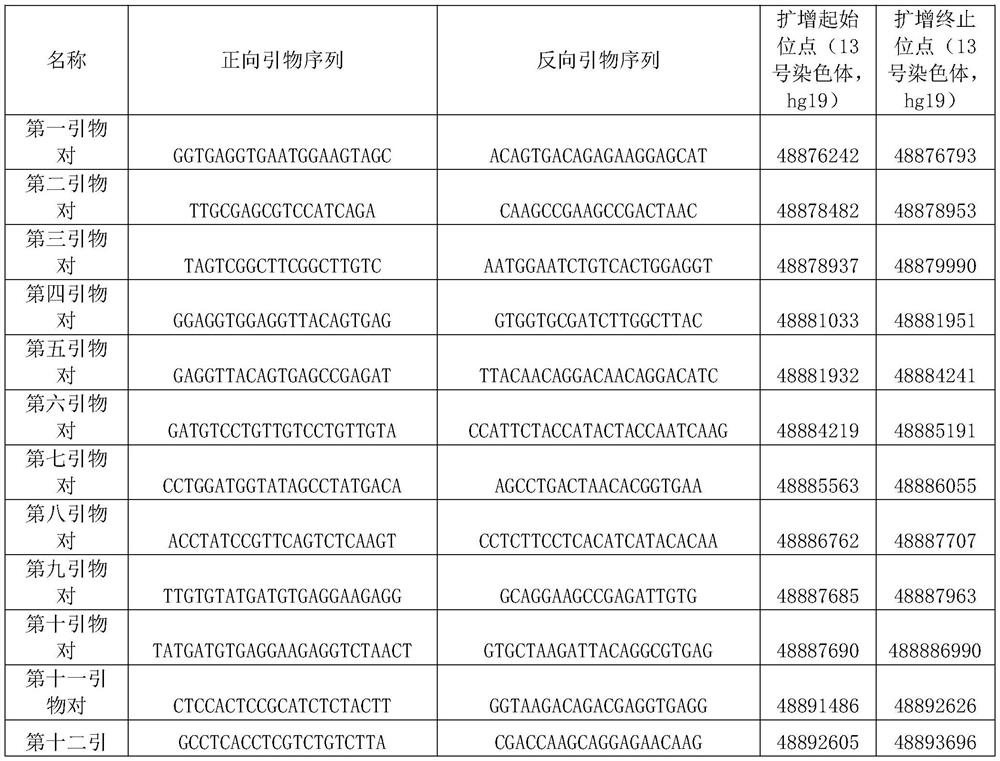
[0027] Genomic DNA was extracted from a human esophageal cancer tissue and matched normal esophageal mucosal epithelium using a conventional spin column method. The PCR primer set designed and optimized by the present invention is used to amplify the genomic DNA gene by PCR. The amplification system was set to 20ul, and each system included 20ng of genomic DNA. After 35 cycles of amplification using a conventional PCR thermal cycler, 5 ul of the product was electrophoresed on a 1.4% agarose gel at a voltage of 160V for 25 minutes. Exposure using an exposure meter, the results of amplification products (such as figure 1 ) for analysis. In the primer pair involved in the present invention, the main band of the amplification product is clear and free of stray bands, and the result is extremely easy to read. After a simple comparison, find the regions with different amplification results in the tumor and normal tissues (red dashed box and green dashed box), then it can be judge...
Embodiment 2
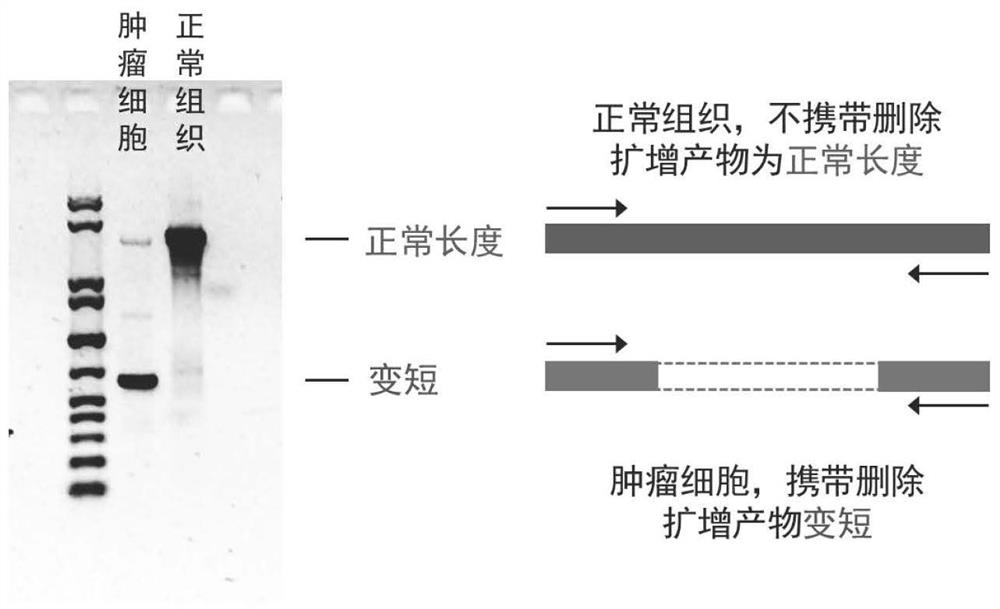
[0029] Genomic DNA was extracted from a human esophageal cancer tissue and matched normal esophageal mucosal epithelium using a conventional spin column method. The PCR primer set designed and optimized by the present invention is used to amplify the genomic DNA gene by PCR. The amplification system was set to 20ul, and each system included 20ng of genomic DNA. After 35 cycles of amplification using a conventional PCR thermal cycler, 5 ul of the product was electrophoresed on a 1.4% agarose gel at a voltage of 160V for 25 minutes. Exposure using an exposure meter, the results of amplification products (such as figure 2 ) for analysis. By comparison, it can be found that the amplification product of tumor cells is shorter than that of normal tissue, suggesting that there is deletion in the amplification range. Depending on the primer pair design, the exon deletion can be located within about 5.0 kb of chromosome 13 49044229-49049241. Subsequent experiments proved that chro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com