D-lactic dehydrogenase SaDLD as well as encoding gene and application thereof
A technology of lactate dehydrogenase and gene, applied in application, genetic engineering, plant genetic improvement, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
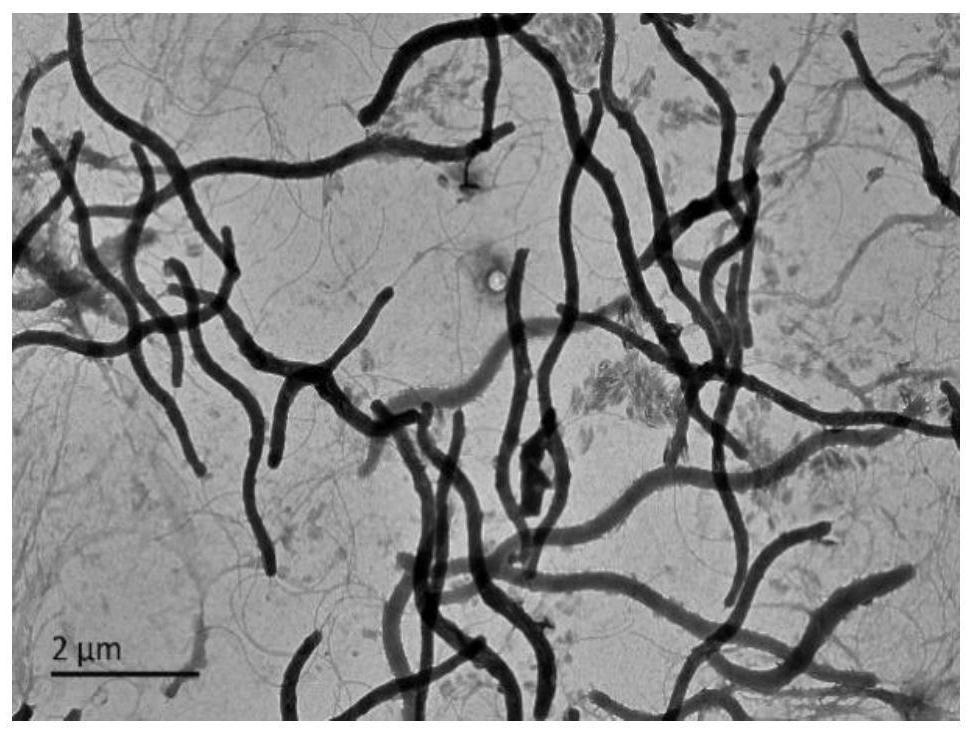
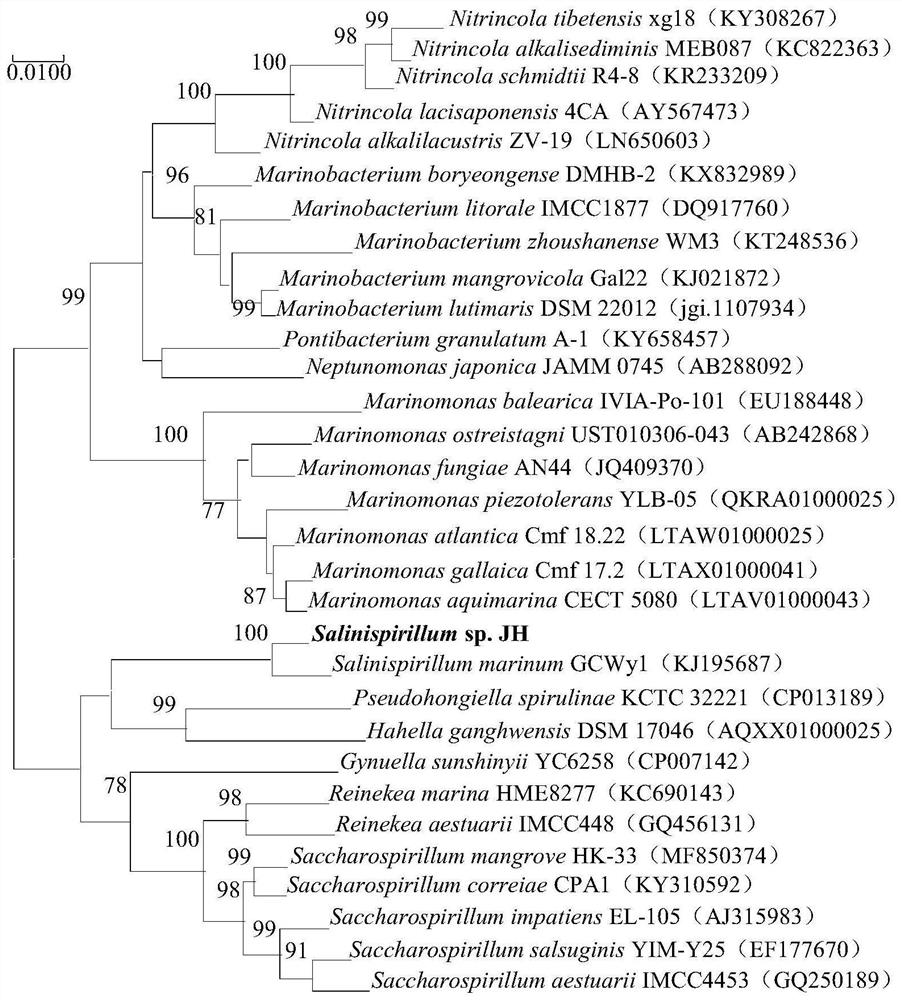
[0019] Example 1, Isolation, Identification and Preservation of Halospiral JH
[0020] 1. Separation
[0021] Take 50 μL of the alkali lake sample, add 450 μL of the alkali lake filtrate, and mix by pipetting to obtain 10 -1 Concentration samples were diluted sequentially to obtain 10 -1 、10 -2 、10 -3 、10 -4 、10 -5 concentrated samples. Take 20 μL, 40 μL of the original solution of the alkali lake water sample and the diluted samples, and spread them on solid medium such as YMAH, TSAH, LBH, 2216EH, etc., culture them in a constant temperature incubator at 35°C for 3-4 days, and observe the growth conditions. Colonies with different colors and shapes were picked, purified and cultivated by the three-section line method, and after repeating the line purification process 3-4 times, pure colonies were obtained.
[0022] 2. Identification
[0023] The purified strains were lined in three zones in LBH solid medium, cultured at 35°C for 2 days, and the morphology, size, prese...
Embodiment 2
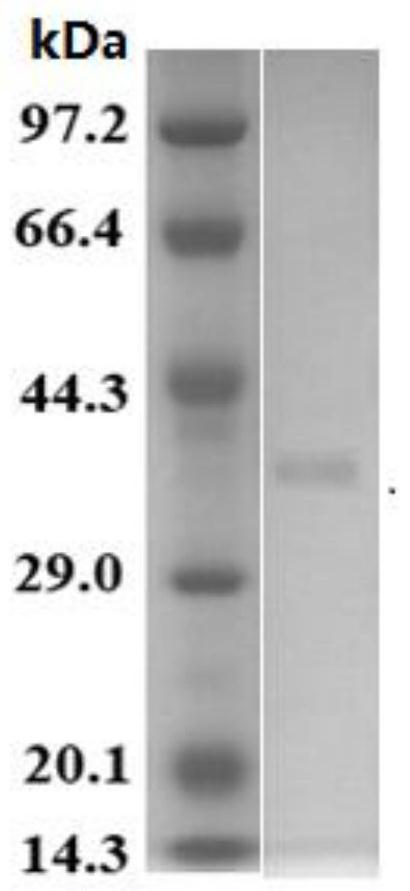
[0034]Embodiment 2, the preparation of D-lactate dehydrogenase (SaDLD protein)
[0035] After a lot of sequence analysis, alignment and functional verification, a new protein was found from Halospira JH, which was named SaDLD protein, as shown in sequence 1 of the sequence listing. The gene encoding SaDLD protein in Halospira JH is named as SaDLD gene, and its coding frame is shown in sequence 2 of the sequence listing.
[0036] 1. Construction of recombinant plasmids
[0037] 1. Using the genomic DNA of Halospiral JH as a template, PCR amplification is performed using a primer pair composed of DL-F and DL-R, and the PCR amplification product is recovered.
[0038] DL-F: 5'-GGAATTCATGAAAATCGCCGTCT-3';
[0039] DL-R: 5'-CCCAAGCTTTTATATCTTAACGACGTGA-3'.
[0040] 2. Take the PCR amplification product obtained in step 1 and connect it with the pET-28a vector to obtain the recombinant plasmid pET-28a-SaDLD.
[0041] pET-28a Vector (pET-28a Vector): Novagen, catalog number 69864...
Embodiment 3
[0054] Embodiment 3, the enzymatic property of D-lactate dehydrogenase (SaDLD protein)
[0055] PBS buffer (100mM, pH 6.0): Weigh 2.88g sodium dihydrogen phosphate, 0.48g potassium dihydrogen phosphate, 0.40g potassium chloride, 16.00g sodium chloride, dissolve in 800mL ultrapure water, adjust pH with HCl to 6.0, set the volume to 1L.
[0056] Sodium alanine reaction solution (20mM): Weigh 2.2g of sodium alanine, dissolve in PBS buffer, and dilute to 1L.
[0057] NADH solution (10 mM): Weigh 6.6343 g of NADH, dissolve in PBS buffer, and dilute to 1 L.
[0058] 1. The effect of pH on the activity of D-lactate dehydrogenase
[0059] 1. Optimal pH
[0060] Get the SaDLD protein solution prepared in Example 2, dilute to 2 times the volume with PBS buffer (100mM, pH 6.0), and use the diluted solution as the test solution.
[0061] Detection method: Add 10 μL of test solution, 10 μL of sodium pyruvate solution (20 mM), 10 μL of NADH solution (10 mM), 170 μL of PBS buffer solutio...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com