CAPS molecular marker for identifying oncidium varieties, screening method and application
A technology of molecular markers and screening methods, applied in the field of molecular biology, can solve problems such as no relevant literature reports, and achieve the effect of making up for the lack of CAPS molecular markers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
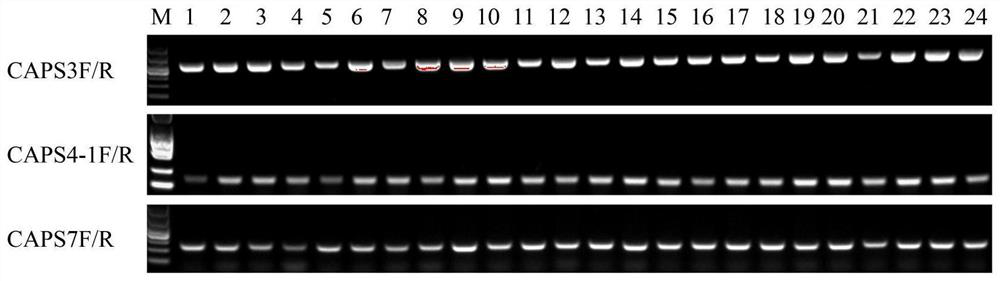
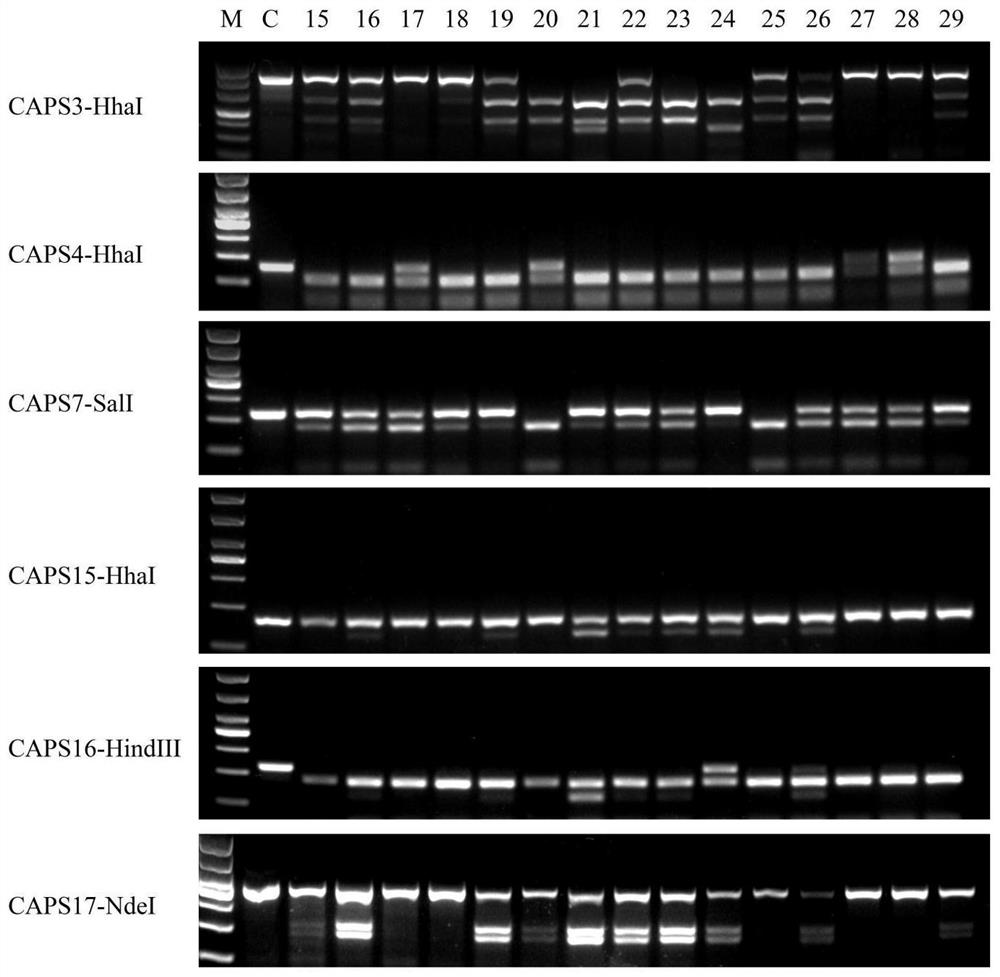
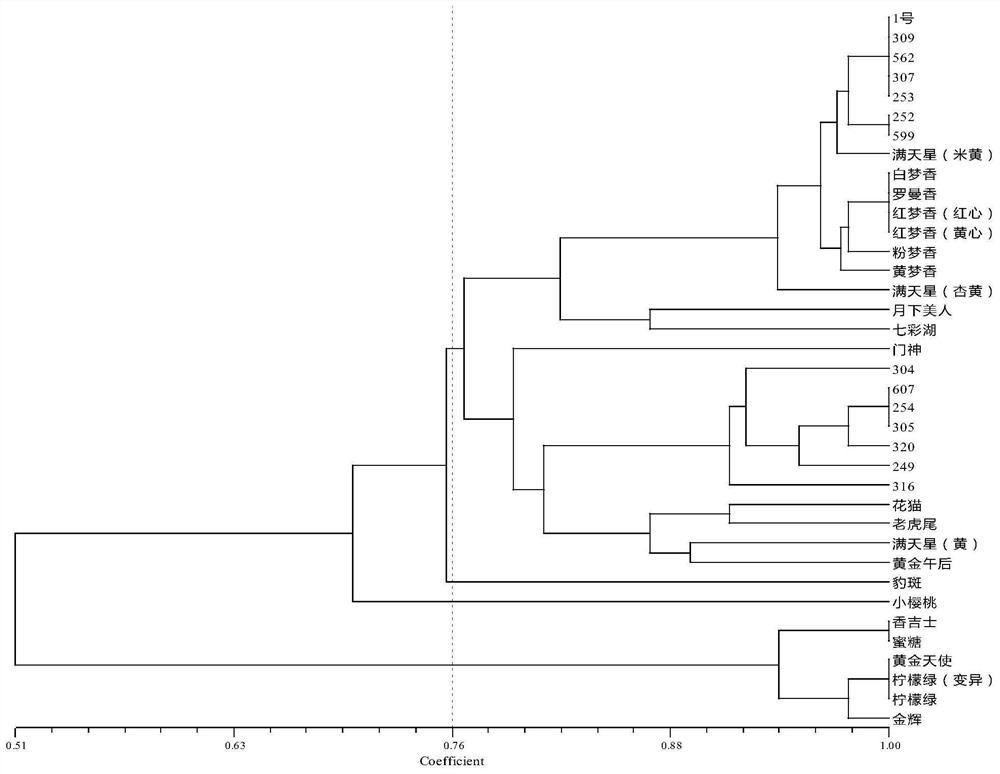
[0046] Step 1 Mining of SNP sites of oncidium cultivars and design of CAPS primers
[0047] (1) Transcriptome sequencing of Oncidium 'Jinhui', 'Perfume', 'Fenmengxiang', 'Hongmengxiang', 'Huangmengxiang' and 'Whitemengxiang'. Using tools such as samtools and picard-tools to compare and compare the results, sort the chromosome coordinates, remove duplicate reads, etc., and finally use the variation detection software GATK2 to perform SNP Calling, and filter the original results (filter out quality values less than 30 and distances less than 5 SNP), get the SNP information.
[0048](2) In the oncidium transcriptome sequence, search for SNP sites in the unigene of genes related to anthocyanin biosynthesis and floral fragrance biosynthesis, and copy the 30 bp sequences upstream and downstream of the SNP site to the online enzyme recognition software dCAPS Find2 .0 (http: / / helix.wustl.edu / dcaps / dcaps.html), looking for SNP sites that can cause changes in the recognition sites of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com