Escherichia coli Nissel 1917 genetically engineered bacterium with hypoglycemic effect, and preparation method and application of escherichia coli Nissel 1917 genetically engineered bacterium
A technology of genetically engineered bacteria and Escherichia coli, applied in the field of genetic engineering, can solve problems such as limiting physiological efficacy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Embodiment 1: Acquisition and identification of Escherichia coli Nissle 1917 engineering bacteria
[0021] 1. Acquisition and identification of Nissle 1917
[0022] The feces of healthy human beings are stored in 50% glycerol, and diluted with sterile phosphate buffered saline (PBS). Do two repetitions. Put the petri dish into a 37°C incubator for aerobic culture for 48 hours, and then select smooth, convex, neat edges, and semipermeable white colonies for microscopic examination. The suspected colonies were isolated and purified again in LB medium according to the above steps until the colonies were completely purified. The purified colony was subcultured for 3 times, and then stored at -80°C with a 1:1 mixture of 50% glycerol and bacterial solution.
[0023] 2. Construction of Escherichia coli Nissle 1917 engineering bacteria
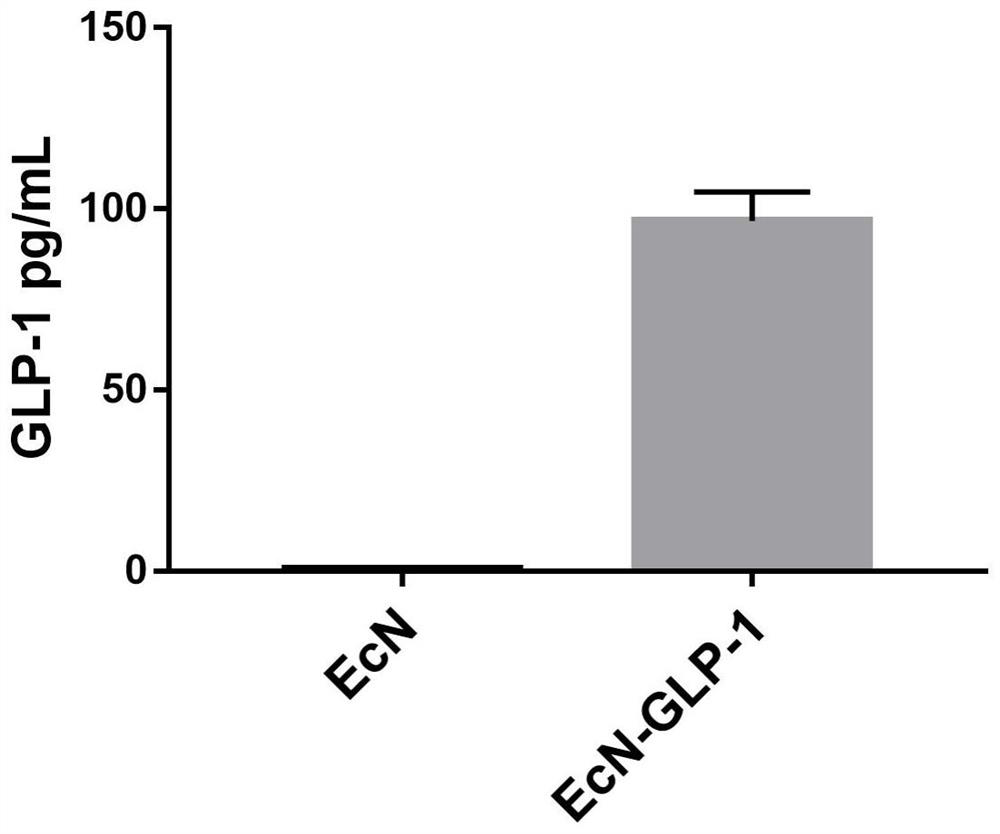
[0024] The GLP-1 target gene was synthesized by chemical synthesis, and the nucleotide sequence was shown in SEQ ID NO.2, and then integra...
Embodiment 2
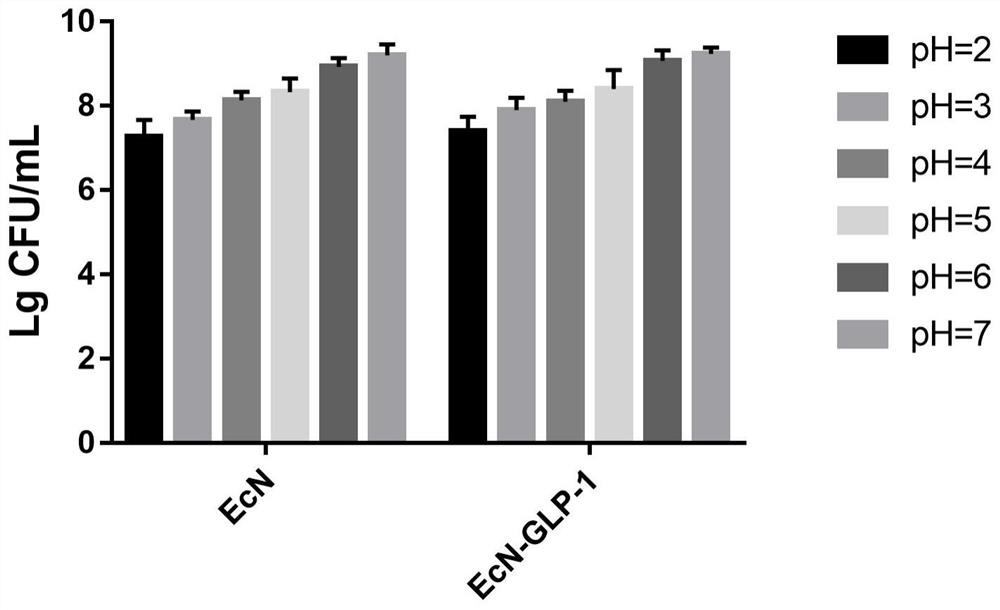
[0051] Embodiment 2: the acid resistance test of Escherichia coli Nissle 1917 engineering bacteria
[0052]Escherichia coli Nissle 1917 and engineering bacteria were inoculated at a volume of 1 / 100, and subcultured in LB liquid medium for 18 hours. Centrifuge at 8000rpm for 5min, retain the precipitate and discard the supernatant, wash twice with sterile PBS buffer, divide each into five 1.5mL centrifuge tubes, and centrifuge at 8000rpm for 5min to obtain wild-type Escherichia coli Nissle 1917 and engineering 5 copies of each bacterial cell. Then add pre-configured PBS buffer solutions with pHs of 2.0, 3.0, 4.0, 5.0, 6.0 and 7.0 respectively, place them in a 37°C incubator and cultivate them for 2 hours. times) dilution, the number of viable bacteria was determined by plate counting method, and the survival rate of wild-type E. coli Nissle 1917 and engineering bacterial strains was calculated.
[0053] The result is as figure 2 As shown, in the acid resistance experiment, ...
Embodiment 3
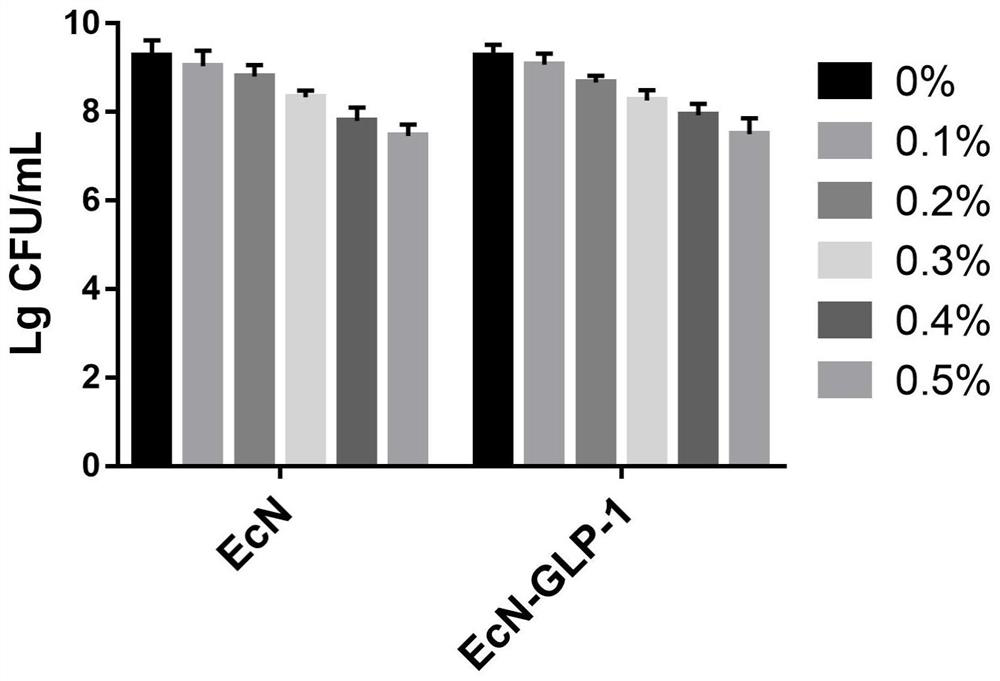
[0054] Example 3 Escherichia coli Nissle 1917 Engineering Bacteria Bile Salt Resistance Experiment
[0055] Escherichia coli Nissle 1917 and engineering bacteria were inoculated at a volume of 1 / 100, and subcultured in LB liquid medium for 18 hours. Centrifuge at 8000rpm for 5min, save the precipitate and discard the supernatant, wash twice with sterile PBS buffer, divide each into five 1.5mL centrifuge tubes, centrifuge at 8000rpm for 5min, and obtain wild-type E. coli Nissle 1917 and engineered bacteria 5 copies of bacteria. Then add pre-configured PBS buffer solutions with bile salt concentrations of 0%, 0.1%, 0.2%, 0.3%, 0.4% and 0.5%, respectively, place them in a 37°C incubator for 2h, and take 0h and After 2h, the bacterial suspension was diluted in multiple ratios (10 times), and the number of viable bacteria was determined by plate counting, and the survival rate of wild-type E. coli Nissle 1917 and engineered strains was calculated.
[0056] The result is as imag...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com