Comparative transcriptome analysis method for peanut leaf gene differential expression under intercropping corn
A technology of transcriptome analysis and gene difference, applied in biochemical equipment and methods, sequence analysis, microbial measurement/inspection, etc., can solve problems such as lack of research, unexplored and analyzed, and impact on dwarf crop productivity, and achieve Method Accurate Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] Comparative Transcriptome Analysis of Peanut Functional Leaf Response to Intercropping Maize Stress at Pod Expansion Stage:
[0068] 1. Establishment of corn intercropping peanut system:
[0069] Peanut variety: Osmanthus 836; this variety is a peanut variety that is mainly planted in Guangxi for disease resistance, high yield and suitable for intercropping;
[0070] Corn variety: Guidan 0810; this variety is a high-yield corn variety in Guangxi;
[0071] Experimental address: Wuming Lijian Base, Guangxi Academy of Agricultural Sciences;
[0072] Peanut planting time: March 10, peanut pod expansion period sampling time: June 15.
[0073] Soil conditions: total nitrogen, total phosphorus and total potassium content are 0.113%, 0.053%, 0.24% respectively, hydrolyzable nitrogen 88mg / kg, available phosphorus 11.4mg / kg, available potassium 110mg / kg, organic matter 18.5g / kg; Red loam, PH value 7.1.
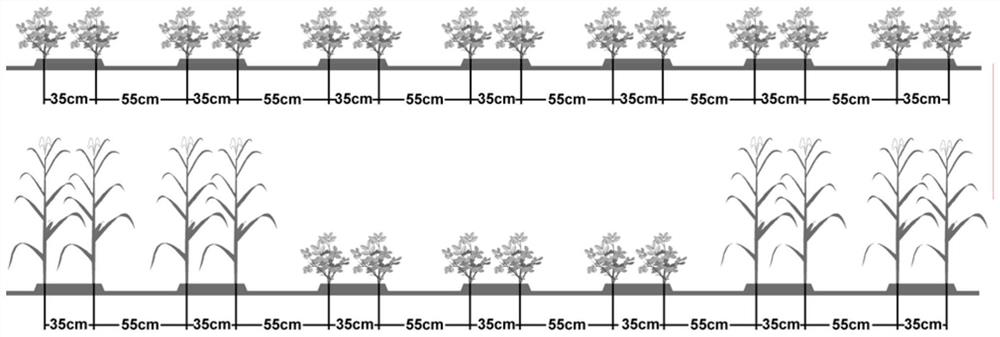
[0074] Experimental design: a randomized block design was adopted, with ...
Embodiment 2
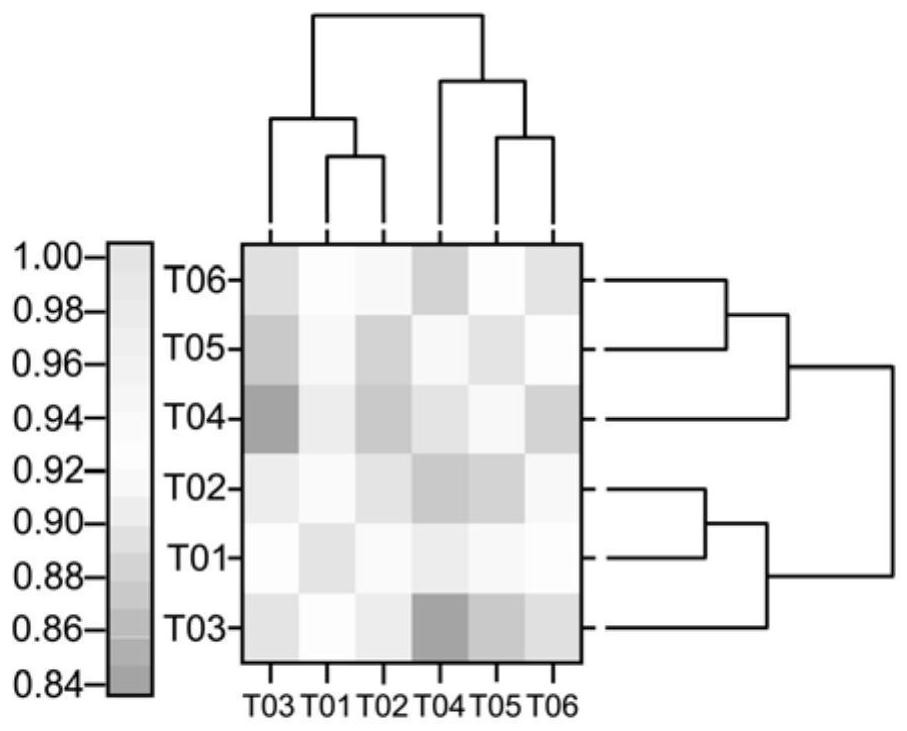
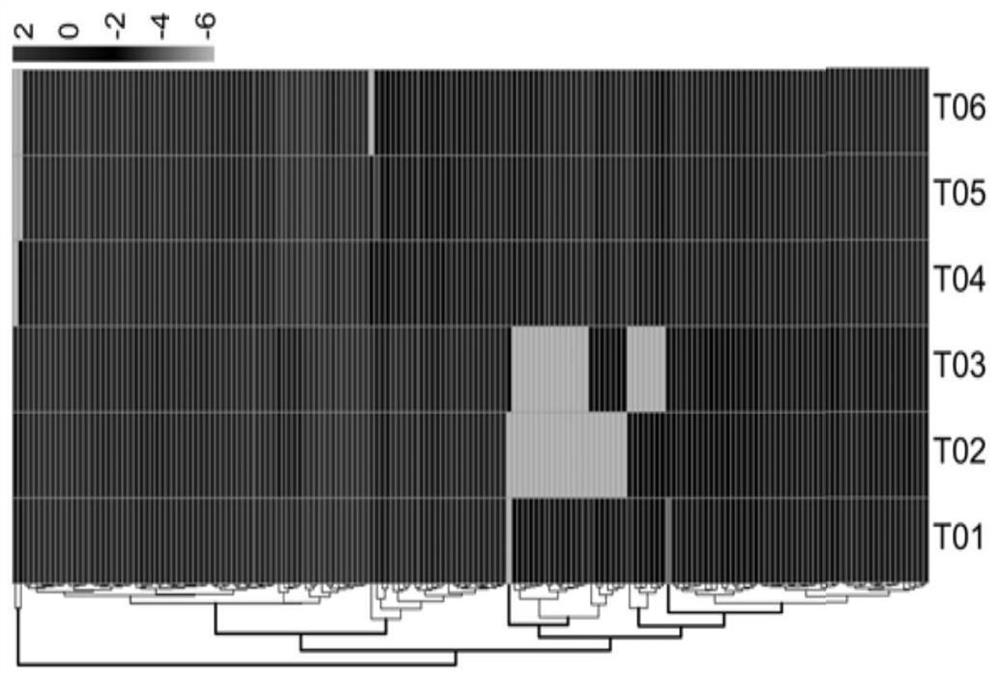
[0145] qRT-PCR verification of differentially expressed genes
[0146] In order to verify the reliability of transcriptome sequencing, 17 differentially expressed genes (DEGs) were selected for real-time fluorescent quantitative PCR test, and the transcriptome sequencing results were compared according to Example 1, and 17 genes were selected to be related to carbon and nitrogen metabolism, amino acid metabolism, Differentially expressed genes related to lipid metabolism, sugar metabolism and chlorophyll metabolism as well as plant hormone signaling and plant-pathogen interactions, plus ribose phosphate-3-epimerase and glycerol-3-phosphate-2-O- For the two important genes of acyltransferase, Actin11 was selected as the internal reference gene. Design specific primers, the primer sequences are shown in Table 8:
[0147] Table 8 Primer pairs for important differentially expressed genes (see SEQ ID NO.1-SEQ ID NO.36 for details)
[0148]
[0149]
[0150] The method for a...
Embodiment 3
[0154] After designing the experiment according to the method of Example 1, in the peanut pod expansion stage, select 3 peanut plants of normal growth from each plot of peanut single cropping (contrast), and from each row of each plot of corn intercropping peanut (treatment) Select 3 normal growing peanut plants (18 plants in total), investigate and measure the plant height and lateral branch length of peanut plants under two planting modes of peanut single cropping and corn intercropping peanut, as well as peanut functional chlorophyll SPAD value and net photosynthetic rate, The chlorophyll SPAD value of the inverted three-leaf functional leaf of the peanut main stem was measured using a Japanese SPAD-502 chlorophyll meter, and the net photosynthetic rate Pn value of the inverted three-leaf functional leaf of the peanut main stem was measured using a CI-340 portable photosynthetic analyzer produced by CID Corporation of the United States Measured during the time period from 9:...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com