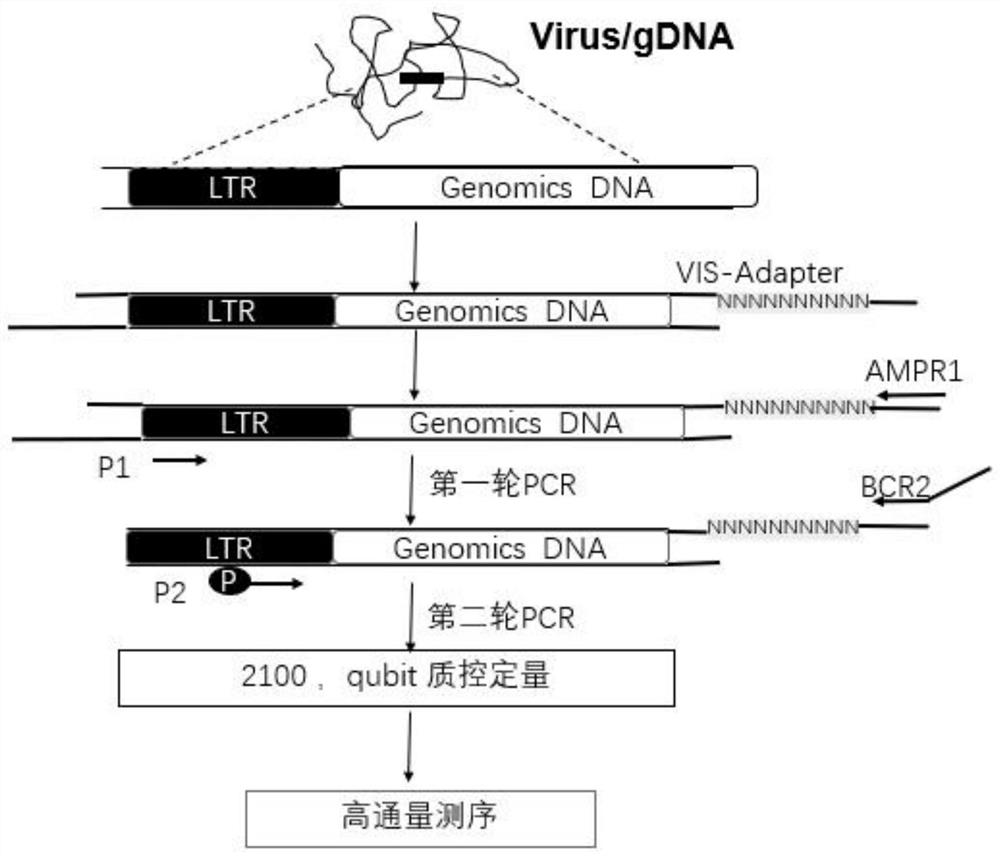
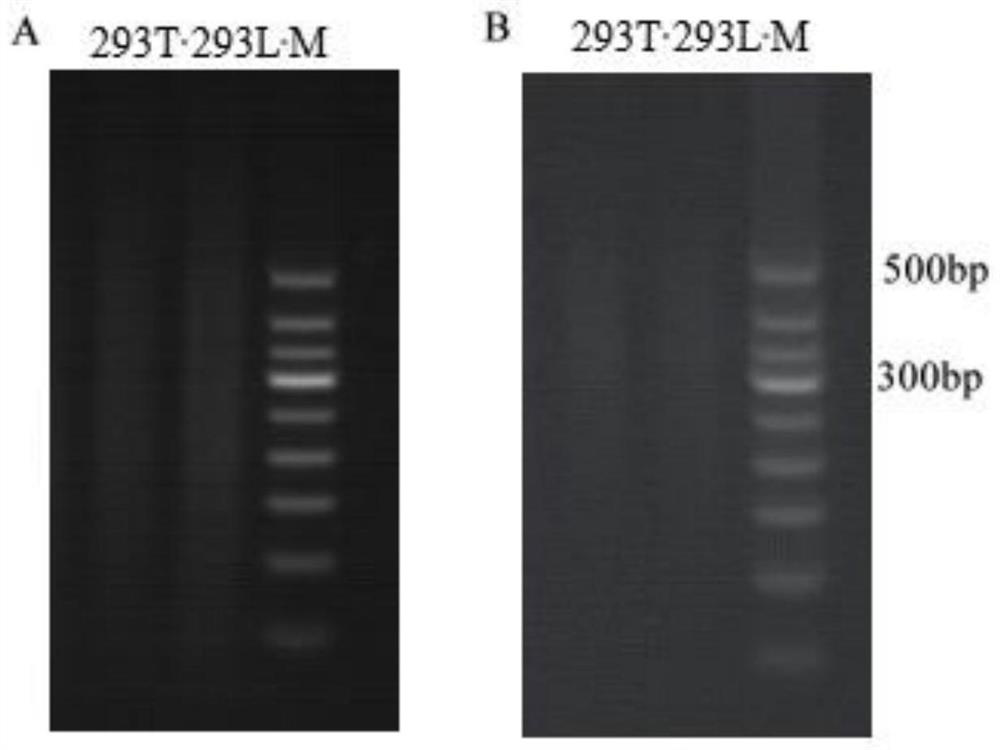
Sequencing library construction method for detecting lentivirus insertion sites, and lentivirus insertion site detection method
A technology of inserting sites and sequencing libraries, which is applied in the field of gene analysis and detection, can solve the problems of low binding efficiency of biotin affinity chromatography, increase of synthesis cost and working time, and high initial amount of template, so as to reduce synthesis cost and purify The effect of cost, low cost, and small initial amount
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] The insertion site analysis of the lentiviral vector in the genome of the 293T cell line comprises the following steps:
[0073] 1. Genomic DNA Extraction
[0074] 1.1 Collect the cells infected with lentivirus, wash them twice with PBS, add 0.05% trypsin to digest for 3 minutes, collect the cells and centrifuge, and discard the supernatant.
[0075] 1.2 Use Tiangen Blood Cell Tissue Genome Extraction Kit for genome extraction.
[0076] 1.3 Use Qubit to measure DNA concentration.
[0077] 2. Random interruption of DNA
[0078] 2.1 Take 100ng~1μg of DNA from the sample for fragmentation. If there is RNA contamination, add 1-2µl RNase for digestion before fragmentation, and quantify to 80-100µl for later use. Ultrasonic instrument CovarisLE210, Dutyfactor (%) 20, Cyelesperburst200, Waterlevel6, Intensity5, 105s.
[0079] 2.2 Use magnetic beads to purify the interrupted fragments, mix with the interrupted product according to the ratio of 0.6 / 0.2, let it stand at room ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com