Sequence-graph based tool for determining variation in short tandem repeat regions
A technology of repetitive sequences and sequence diagrams, which can be used in sequence analysis, genomics, instruments, etc., and can solve problems such as difficulty in determining repetitive sequences.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
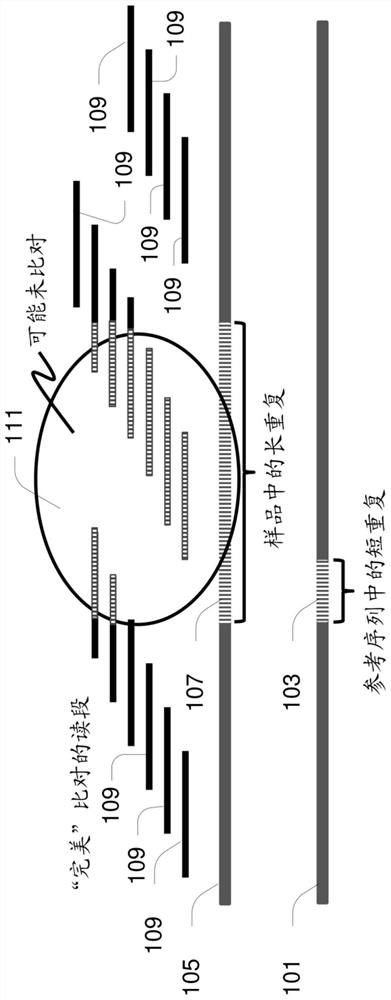
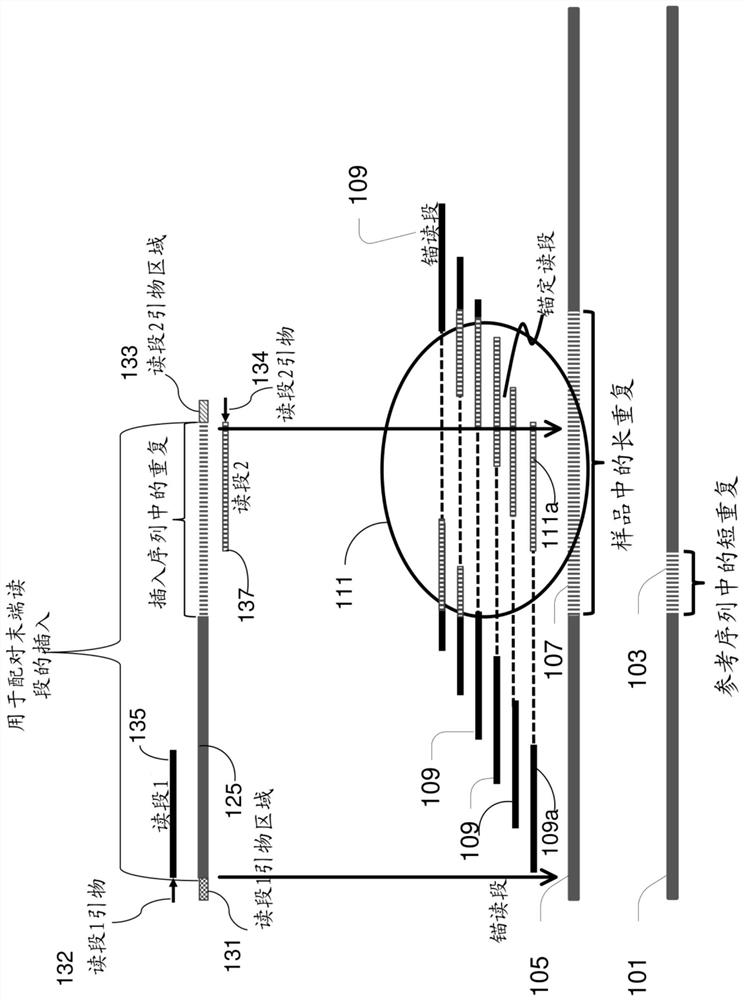
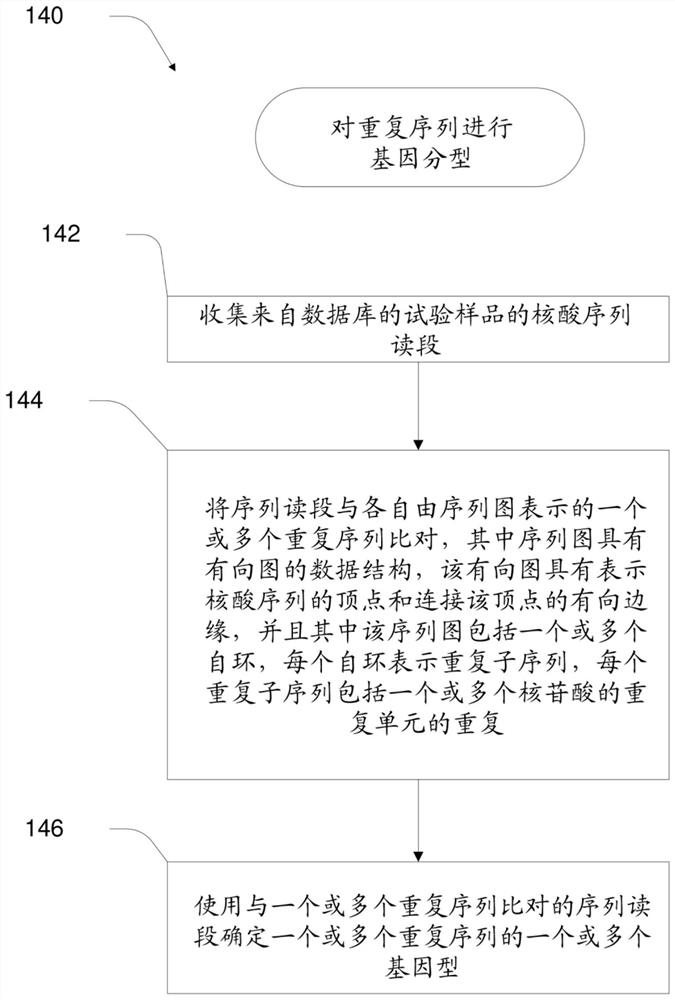
[0042] The present disclosure relates to methods, apparatus, systems and computer program products for identifying repeat expansions of interest, such as expansions of medically significant repeat sequences. Examples of repeat expansions include, but are not limited to, those associated with genetic disorders such as fragile X syndrome, ALS, Huntington's disease, Friedrich's ataxia, spinocerebellar ataxia, spinobulbar muscular atrophy, myotonic Amplifications associated with dystrophy, Machado-Joseph disease, and dentatoerythropallidal Lewy body atrophy).
[0043] Unless otherwise indicated, the practice of the methods and systems disclosed herein involves conventional techniques and apparatus commonly used in the fields of molecular biology, microbiology, protein purification, protein engineering, protein and DNA sequencing, and recombinant DNA, which are described herein within the technical scope of the field. Such techniques and devices are known to those skilled in the a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com