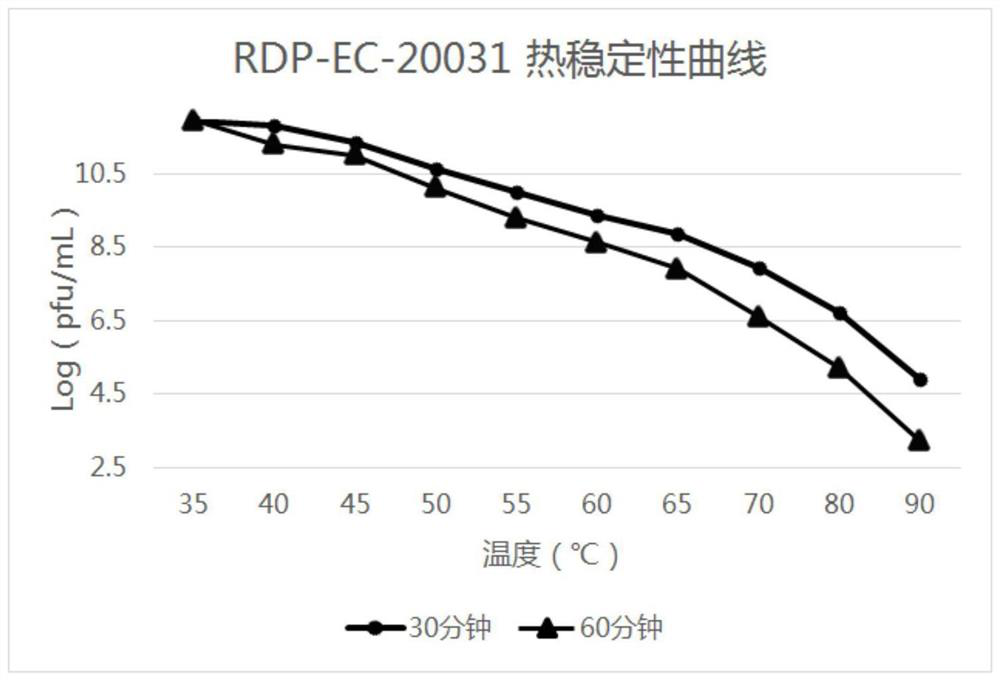
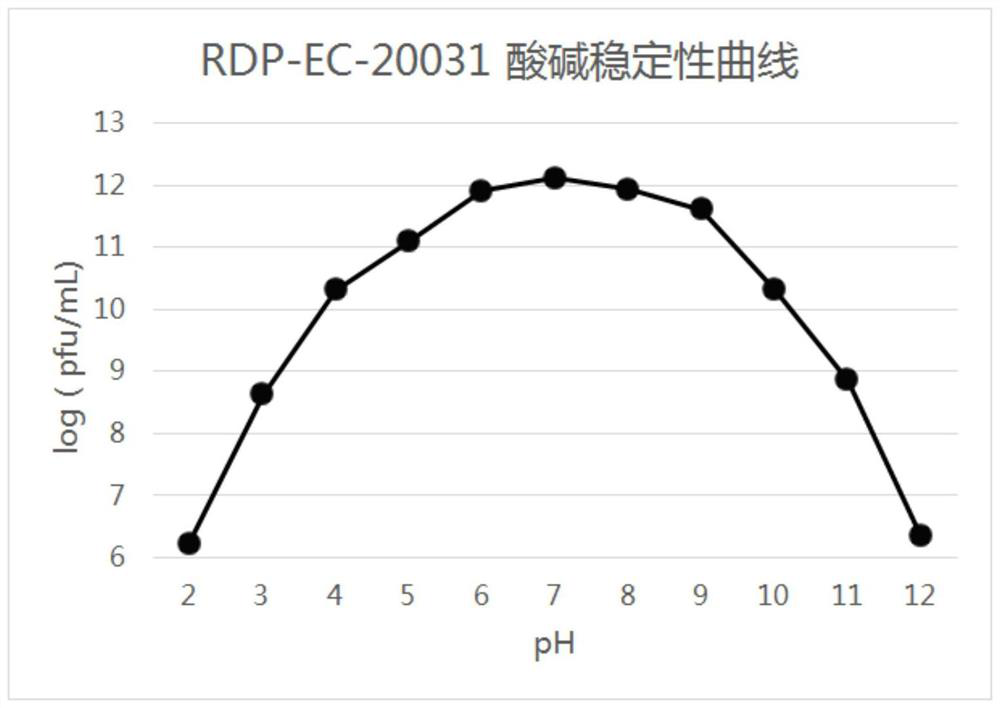
High-temperature-resistant Escherichia coli phage RDP-EC-20031 and application of high-temperature-resistant Escherichia coli phage RDP-EC-20031
A technology of RDP-EC-20031 and Escherichia coli, which is applied in the direction of viruses/phages, medical raw materials derived from viruses/phages, applications, etc., can solve the problems of drug resistance of pathogenic bacteria, ineffective medication, and untimely treatment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 Isolation and Identification of Pathogenic Escherichia coli BE-20036
[0035] Sampling from the diseased chicken farm, aseptically take the liver of the diseased chicken, streak it on the selective medium (MacConkey agar medium), cultivate it at 37°C for 18-24 hours, and form a round, flat, edge on the medium For red colonies with neat, smooth and moist surface, pick typical colonies and continue to streak and purify for 3 times, then pick a single colony and inoculate in 5mL LB broth, shake and culture at 37°C and 200rpm for 8h to obtain a uniform turbid bacterial suspension. Then it was identified as Escherichia coli through 16sRNA molecular identification, and its serotype was further determined to be O78 through serum identification. O78 has strong pathogenicity, and can be transmitted to humans through food chains and other methods, and has strong pathogenic infectivity; Name it BE-20036 and store it in a -80°C refrigerator.
Embodiment 2
[0036] Example 2 Isolation and identification of coliphage RDP-EC-20031
[0037] (1) Sewage treatment: take sewage from the farm, centrifuge at 10,000 rpm for 5 minutes, take the supernatant and pass it through a 0.22 μm filter, and collect the filtered supernatant for later use.
[0038] (2) Preparation of mixed bacterial suspension: Take 0.2mL of bacterial suspension and 0.3mL of filtrate and add it to 5mL of LB broth, culture at 37°C, shake at 200rpm overnight, then centrifuge at 10,000rpm for 5min, take the supernatant and pass it through a 0.22μm filter. Set aside the filtrate.
[0039] (3) Separation of bacteriophages: Separation of phages was carried out using the double-plate method. Take 0.2 mL of the filtrate of the mixed bacterial suspension and 0.2 mL of the Escherichia coli suspension and mix evenly, bathe in water at 37°C for 10 minutes, then spread double plates, and incubate at 37°C for 8 After -12h, observe the result. If there are phages, there will be clea...
Embodiment 3
[0042] Morphological observation of embodiment 3 phage
[0043] Take 20 μL of the liquid containing crude phage particles and drop them on the copper grid, let it settle naturally for 15 minutes, and absorb the excess liquid from the side with filter paper, add a drop of 2% phosphotungstic acid (PTA) on the copper grid to stain the phage for 10 minutes, and then use the filter paper to remove the The staining solution was sucked off from the side, and the morphology of the phage was observed with an electron microscope after the sample was dried. The result is as figure 1 shown.
[0044] Depend on figure 1 It can be seen that the bacteriophage RDP-EC-20031 has a polyhedral stereosymmetric head, wrapped in nucleic acid, with a diameter of about 70nm, a tail with a length of about 90nm, a tail sheath, and a neck connecting the head and the tail. According to the Ninth Report of the International Virus Taxonomy Organization Virus Classification, the phage can be classified as ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com