Klebsiella pneumoniae phage P560, phage depolymerizing enzyme Depo43 and application
A technology of Klebsiella and phage, applied in the direction of phage, virus/phage, application, etc., can solve the problem that the actual action spectrum of phage depolymerase is not disclosed, and achieve significant clinical effects and efficient inhibitory effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
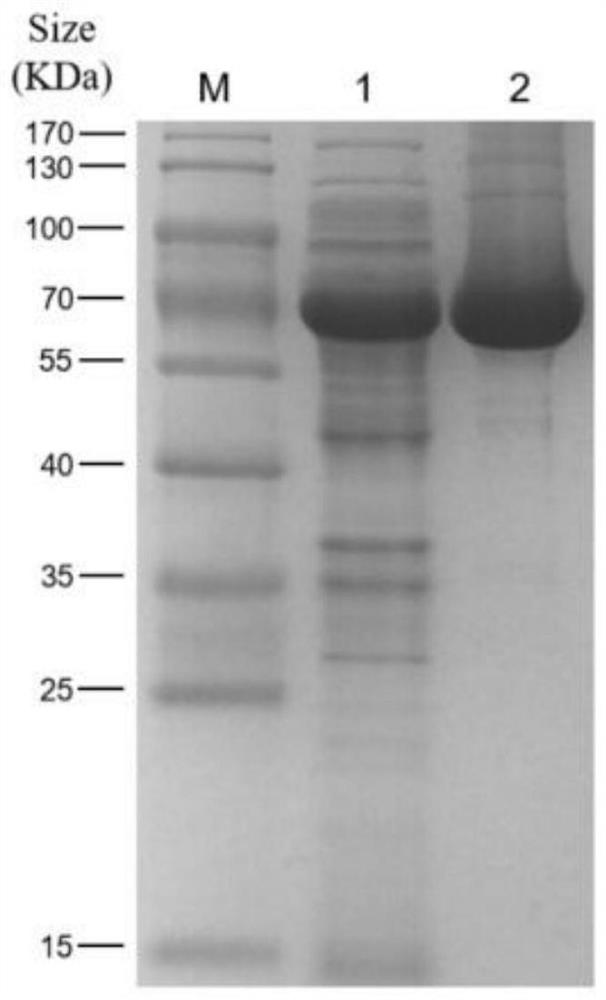
[0026] Example 1 Preparation of phage depolymerase Depo43
[0027] (1) Target product amplification
[0028] Primers designed for constructing depolymerase expression vector:
[0029] The upstream primer F (SEQ ID No: 3) is:
[0030] 5'-GGAATTCCATATGTTAAAACAATCTGAATCAG-3';
[0031] The downstream primer R (SEQ ID No: 4) is:
[0032] 5'-CCGCTCGAGTTATGGACCAATGACCACACC-3'.
[0033] The PCR amplification program was denaturation at 95°C for 3 min; denaturation at 95°C for 15 s, annealing at 50°C for 15 s, extension at 72°C for 2 min, and 30 cycles.
[0034] The depo43 gene PCR product from the 43rd open reading frame of Klebsiella pneumoniae phage P560 was digested and purified and ligated into the pET28a plasmid: restriction sites Nde I and Xho I;
[0035] (2) Add the pET28a-depo43 plasmid to 100 μL Escherichia coli BL21 (DE3) competent cells, flick the tube wall to mix, and let stand on ice for 30 minutes. After heat-shocking in a water bath at 42°C for 45 sec, quickly pla...
Embodiment 2
[0046] Example 2 Using phage depolymerase Depo43 to remove K47 and K64 type bacterial capsular polysaccharides respectively
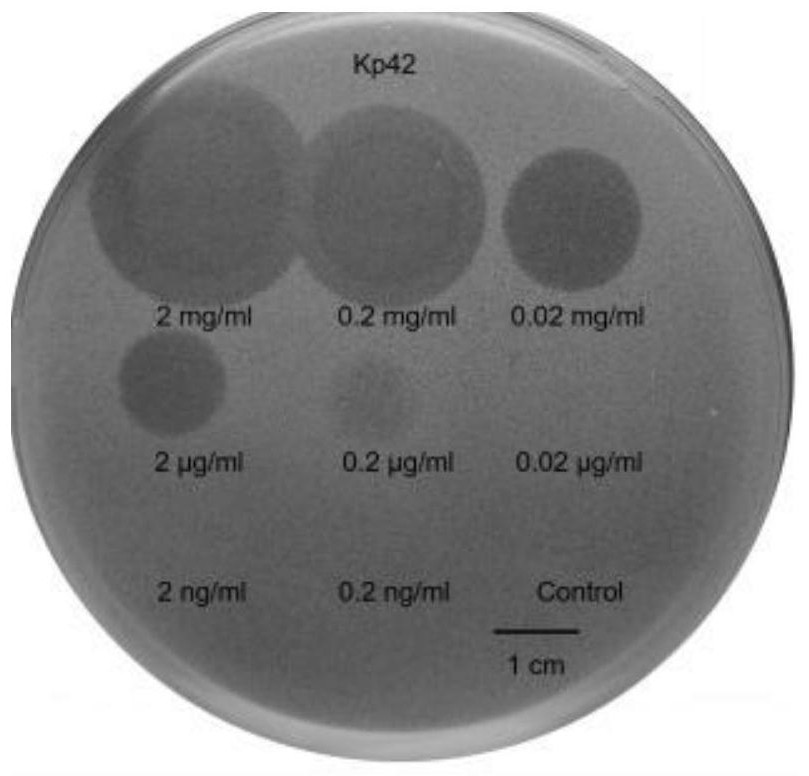
[0047] Use the dot test to detect the inhibitory activity of phage depolymerase Depo43 on E. coli biofilm. Spread 0.5% semi-solid LB medium containing 100 μL of logarithmic bacteria evenly on the solid LB medium, and dry it naturally. A series of diluted concentrations of Dpo43 (2 mg / mL, 0.2 mg / mL, 0.02 mg / mL, 2 μg / mL, 0.2 μg / mL, 0.02 μg / mL, 2 ng / mL and 0.2 ng / mL) were dropped on the double layer plate , the buffer was used as a negative control, cultured overnight in a 37°C incubator, and the phage depolymerase Depo43 with a concentration of 2 mg / mL-0.2 μg / mL formed translucent spots on the plate, such as image 3 shown.
[0048] The extracted capsular polysaccharide was added to a 10% gel (final concentration: 0.3%). After solidification, a 7 mm hole was punched into the gel, and then 20 μL of purified protein and buffer (negative control) were added...
Embodiment 3
[0049] Example 3 Utilizes purified depolymerase depo43 to inhibit K47 type Klebsiella pneumoniae biofilm
[0050] K47 Klebsiella pneumoniae Kp57 80μL (OD 0.4~2×10 8 CFU / mL) were inoculated into a 96-well cell culture plate (Sigma-Aldrich, USA) containing 100 μL LB medium per well. Dilute the purified depo43 with protein buffer to 200 μg / mL, add 20 μL depo43 dilution to each well of the 96-well cell culture plate containing the biofilm, the content is 4 μg, 0.4 μg, 0.04 μg, 4ng, 0.4ng, 0.04 ng and 0.004ng. After culturing for 24 hours, the cell culture plate was taken out, the medium was discarded with a pipette gun and washed twice with sterilized PBS buffer to remove free bacteria and enzymes. Then add 200 μL methanol to each well and fix for 10 minutes. After the fixative was discarded and allowed to dry naturally, the biofilm was stained with 200 μL of 0.1% crystal violet (purchased from Zhuhai Beisuo Biotechnology Co., Ltd.) for 10 minutes at room temperature. After st...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com