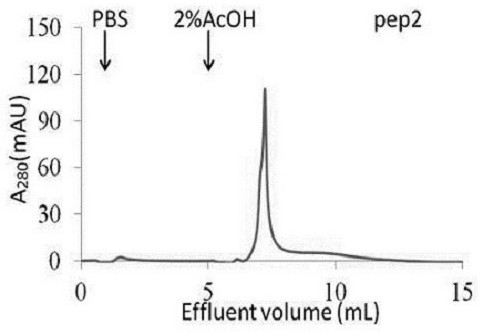
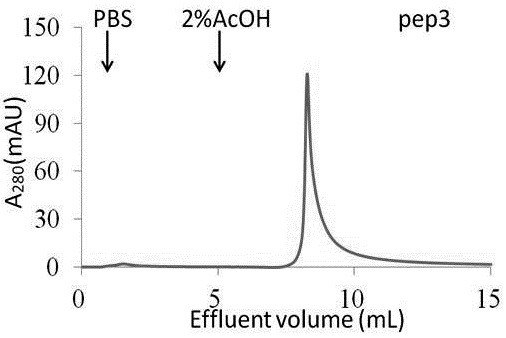
The prognosis prediction model can be used for purifying hydrophobic cyclic peptide ligands of human immunoglobulin G
A human immunoglobulin, hydrophobic technology, applied in the purification of human immunoglobulin G, computer simulation and downstream protein separation and purification, which can solve the problems of low protein purity and difficult commercial utilization.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 Build a simulation system
[0035] Select the crystal conformation of the SpA-hIgG1 complex whose ID is 1FC2 from the PBD database, and extract the C on the SpA and hIgG-Fc fragments H 3 The binding region, the amino acid sequence of the binding region is Ser383-Asn389 (SNGQPEN), and the three-dimensional coordinates of each amino acid residue in the binding region are selected as the coordinate information of the simulation system.
Embodiment 2
[0036] Example 2 Obtaining a Biomimetic Peptide Library
[0037] Use NAMD software to calculate the distance between each residue in the Ser383-Asn389 sequence, take Cys as the connecting residue, insert 5 to 7 amino acid residues X around Cys, and form a circular polytitanium chain-[XXX-C-XXXX ]-, -[XX-C-XXXX]-, -[XX-C-XXXXX]-, -[XXX-C-XXX]-, -[XX-C-XXX]-, using AutoDock molecular docking program for amino acid Positioning, determine the type of amino acid residue X that matches the Ser383-Asn389 coordinate system and has the best binding position. X represents 19 common amino acid residues except Cys, which can be stably combined with hIgG-Fc by calling the GROMOS program The end-to-end biomimetic peptide library of cyclic 6-peptide, cyclic 7-peptide and cyclic 8-peptide sequences, which contains a total of 4256 sequences.
Embodiment 3
[0038] Example 3 Primary Screening of Polypeptide Library
[0039] The peptide molecules in the peptide library were sequentially docked with the three-dimensional structure of hIgG-Fc using LeDock molecular docking software. The docking results showed that most of the peptides could bind to the Fc fragment, and the binding affinity ranged from -3.5 to -7.6kcal / mol Among them, considering the size of the affinity and the screening efficiency, the polypeptide molecules (a total of 1135) with a binding affinity lower than -6.0kcal / mol (absolute value of affinity greater than 6.0kcal / mol) were finally selected for the next round of screening.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com