N-acylhomoserinelactone degrading enzyme and application thereof
An acyl homoserine lactone and enzyme-degrading technology, which is applied in the field of biological enzymes, can solve the problem of low inhibitory activity, and achieve the effect of inhibiting pathogenicity, good thermal stability and pH stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 2
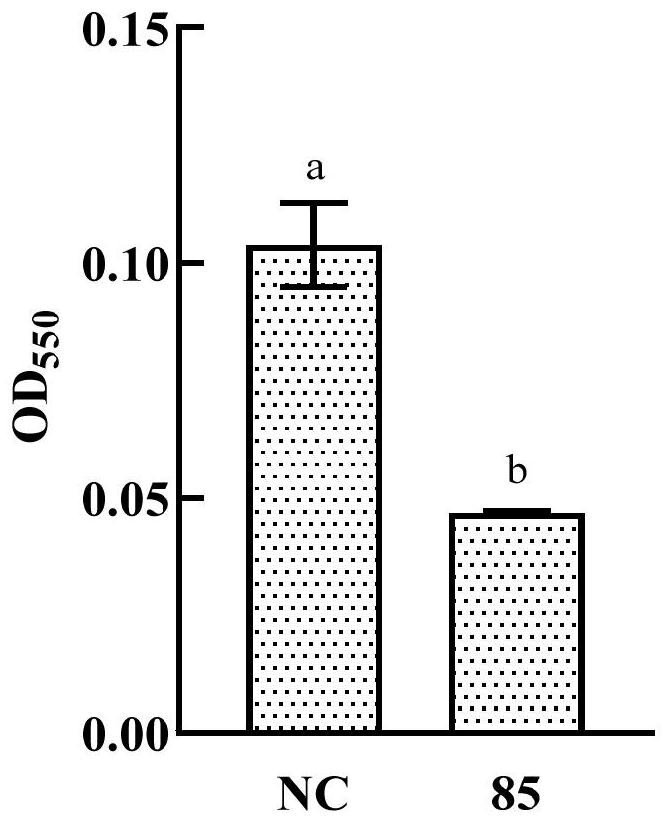
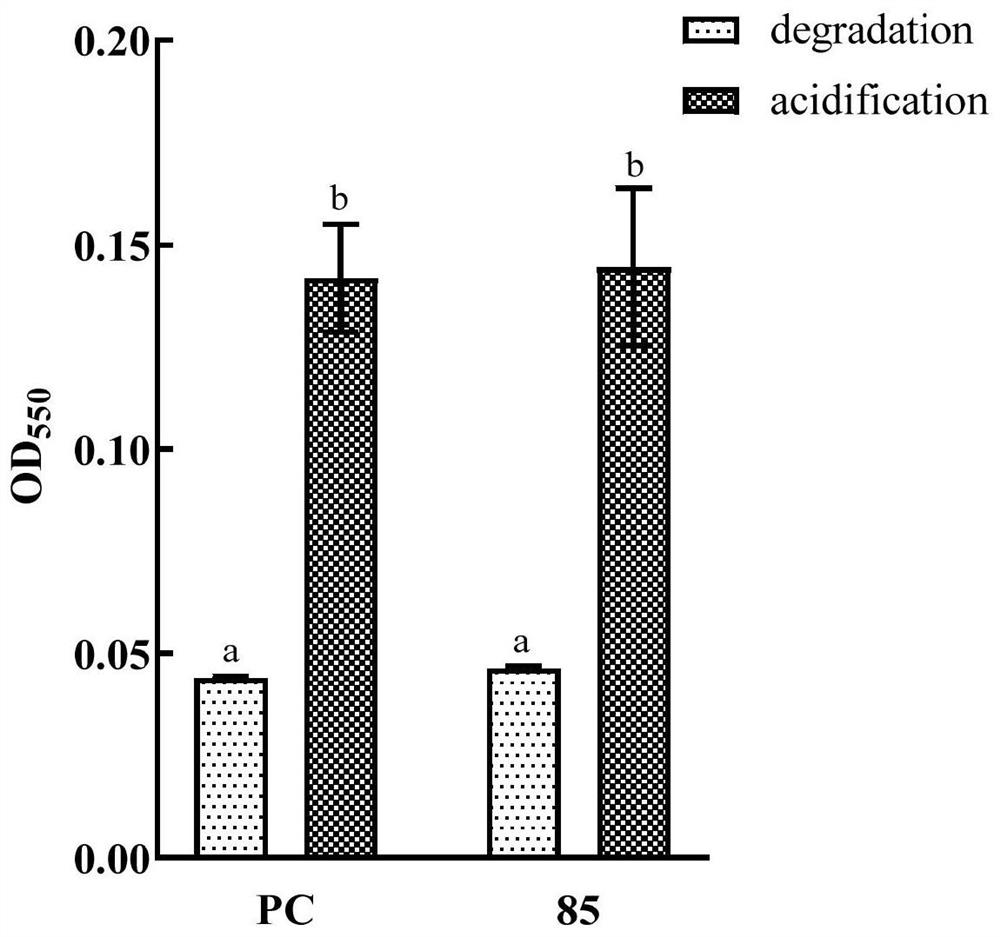
[0044] Example 2 AHL degradation product acidification recovery experiment
[0045] Add the purchased self-inducing molecule (C6-HSL, the structure is ), after shaking for 1 d, centrifuge to get the supernatant. Adjust the pH of the supernatant to 1 with hydrochloric acid, incubate for 1 day under acidic conditions, then adjust the supernatant back to neutral with sodium hydroxide, detect AHL in the supernatant with Chromobacterium violaceum CV026; CV026 produced purple pigment, indicating that the product of the self-inducing molecule degraded by XY-85 bacterial fluid recovered under acidic conditions, which indicated that the AHL degrading enzyme produced by XY-85 was a lactonase.
[0046] The bacterial liquid was centrifuged, the supernatant was removed, and 100 μL of DMSO was added to the bacterial cell to extract the purple pigment in the bacterial cell. Centrifuge again, take 100 μL of the supernatant and add it to the microplate plate, and measure the OD with a micro...
Embodiment 3X
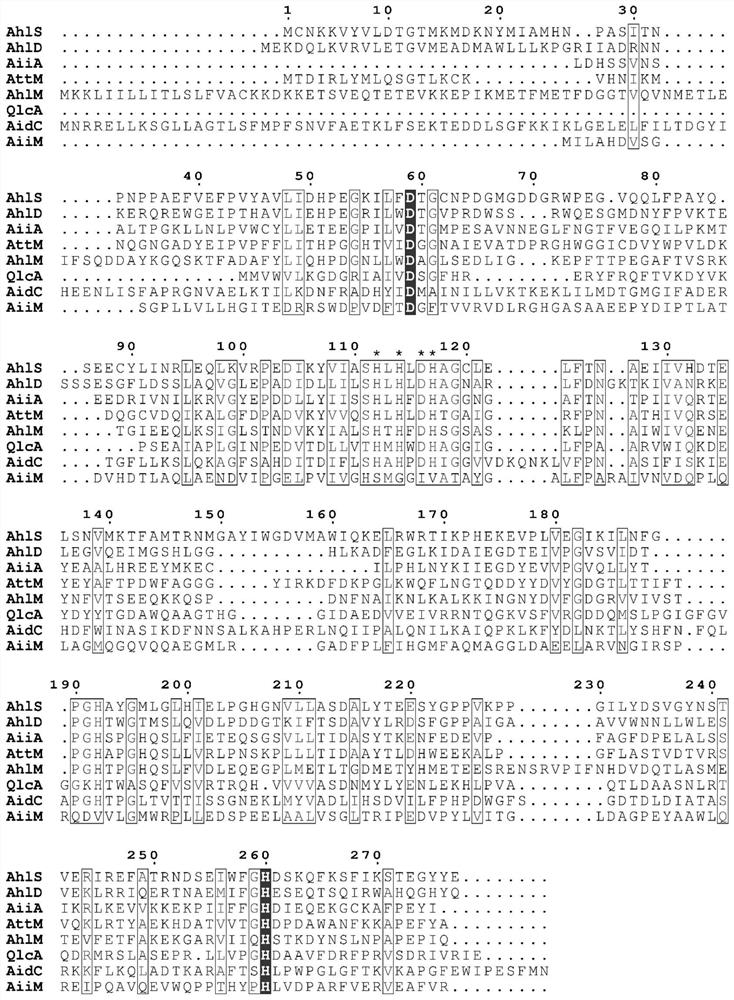
[0050] Embodiment 3XY-85 lactonase sequence analysis
[0051] Through whole genome sequence analysis, the whole genome sequence information of XY-85 was obtained, and the sequence information was compared with the reported lactonase sequence in the KEGG database, and finally XY-85 lactonase (that is, AHL degrading enzyme) was screened Amino acid sequence information (SEQ NO.2), and named AhlM. The amino acid sequence information of AhlM was compared with the amino acid sequences of some reported lactonases by clustalW, and the comparison results are as follows image 3 As shown, it shows that AhlM has conserved amino acids D (No. 59) and H (No. 260), and also contains a conserved region (amino acid with *) between amino acids No. 110 and No. 120.
[0052] In order to explore the evolutionary relationship between AhlM and the reported lactonases, the maximum likelihood method was used to construct the following Figure 4 The AhlM protein sequence phylogenetic tree shown shows...
Embodiment 4
[0053] Example 4 Cloning of the gene AhlM of XY-85 lactonase into Escherichia coli for expression and verification of its AHL degradation activity
[0054] Activate XY-85 into the MA culture plate, culture it upside down at 28°C for 3 days, and then use Genomic DNA was extracted with the SPIN Kit for Soil kit, and the DNA concentration and purity were determined with a nanodrop 2000C. Use the genomic DNA of XY-85 as template, use primer 85NH-F (nucleotide sequence is SEQ NO.3) and 85NH-R (nucleotide sequence is SEQ NO.4) PCR amplification (PCR system sees following (4.1), see the following (4.2) for the PCR program) AhlM (the nucleotide sequence is SEQ NO.2).
[0055] (4.1) PCR system:
[0056] PrimeSTAR Max DNA Polymerase 25 μL;
[0057]
[0058]
[0059] (4.2) PCR program:
[0060]
[0061] The obtained amplified fragment and the pET28a vector were double-digested with Nco I and Xho I endonucleases at 37°C (enzyme digestion system and restriction procedures are...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com