GRNA for targeted destruction of SARS-CoV-2 genome and application of gRNA
A virus genome, sars-cov-2 technology, applied in the field of gene editing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Reporter carrier design and construction.
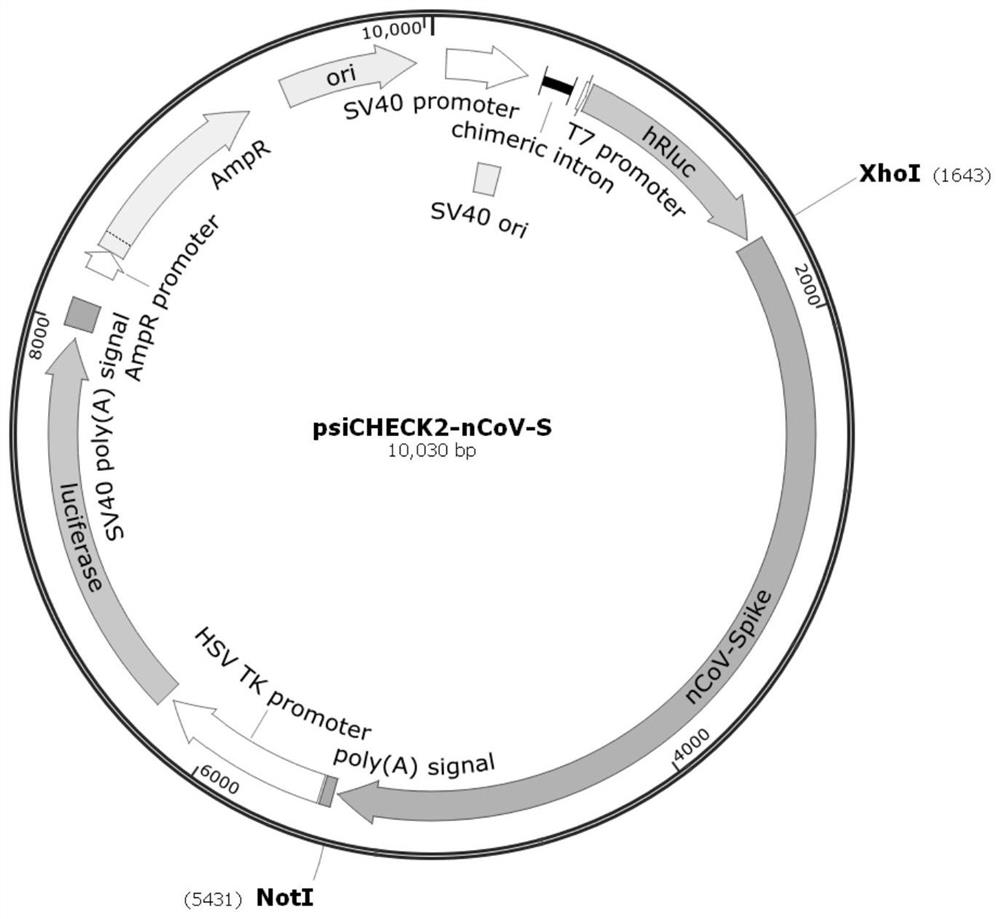
[0055] The psiCHECK2 plasmid was linearized with XhoI and NotI double restriction enzymes, and reacted at 37°C for 1 hour. The digested product was subjected to 1% agarose gel electrophoresis, and the 6242bp digested product was recovered by cutting the gel, and stored at -20°C for future use.
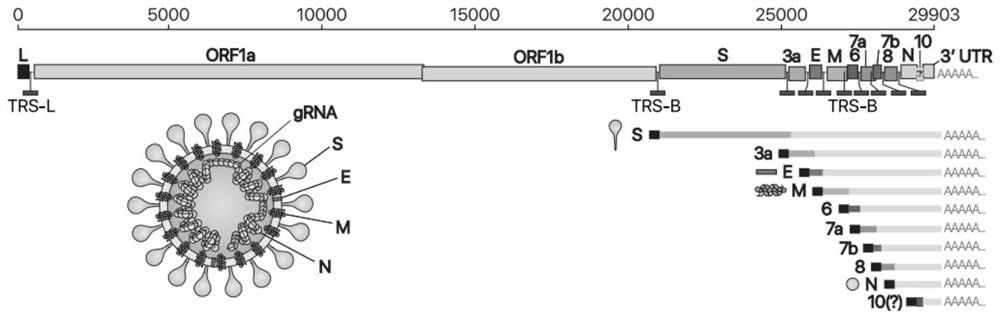
[0056] According to the published SARS-CoV-2 genome sequence (NC_045512.2), its S region was synthesized into the pcDNA6B vector. SARS-CoV-2 genome structure such as figure 1 shown.
[0057] Use PrimeSTAR Max DNA polymerase premix to perform PCR amplification of bases 24-3804bp in the Spike region, and at the same time, a 15-20bp homology arm is introduced into the primers for subsequent cloning between the NotI and XhoI restriction sites of psiCHECK2 , as the 3'UTR of hRluc. The PCR products were subjected to 1% agarose gel electrophoresis, and the target bands were recovered by cutting the gel.
[0058] The PCR recovery product ...
Embodiment 2
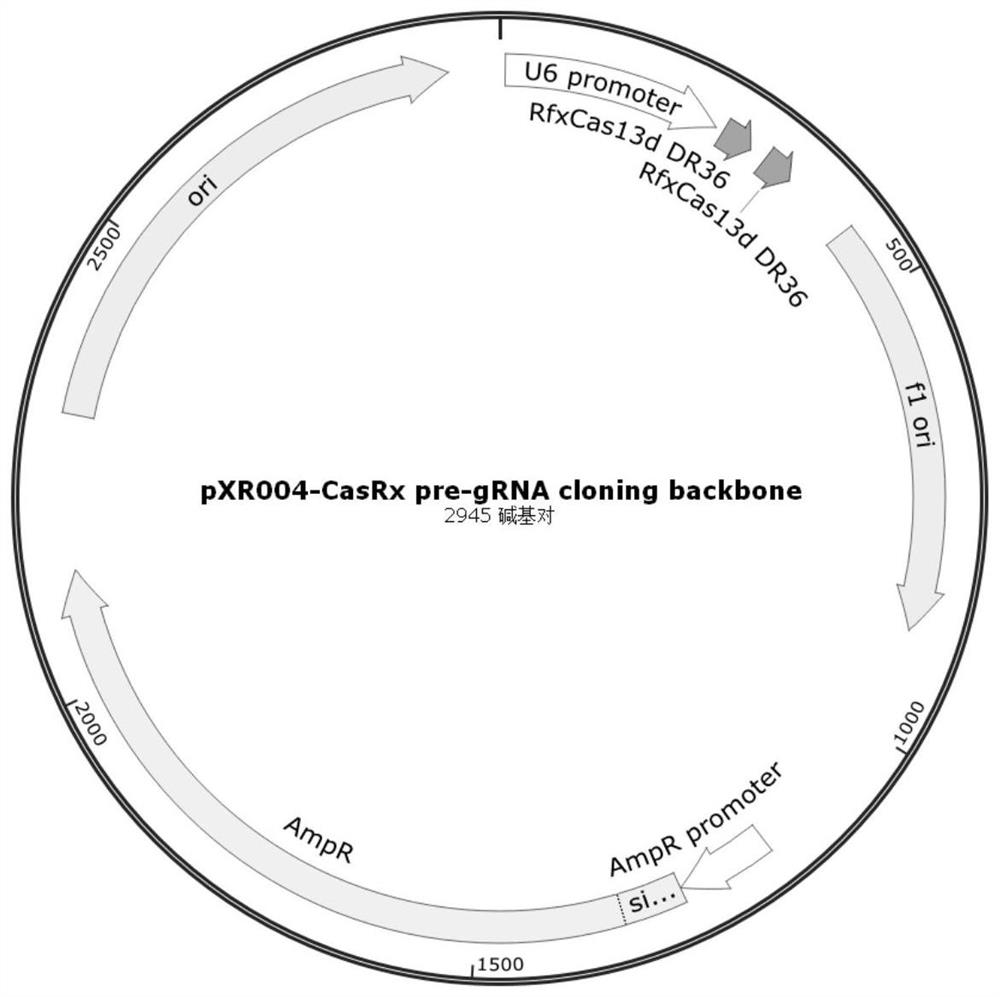
[0063] gRNA vector design and construction.
[0064] A 30nt gRNA targeting domain sequence was designed according to the target sequence in the S region of the SARS-CoV-2 genome, and two gRNAs were designed for the hRluc region as a control. The targeting domains of the gRNA are shown in Table 1.
[0065] Table 1. Targeting domains of gRNAs
[0066] name targeting domain serial number nCoV-S-1 AUAACCCACAUAAUAAGCUGCAGCACCAGC SEQ ID NO:1 nCoV-S-2 UUAGAAUUCCAAGCUAUAACGCAGCCUGUA SEQ ID NO:2 nCoV-S-3 UAGAACCUGUAGAAUAAACACGCCAAGUAG SEQ ID NO:3 nCoV-S-4 UCACCAUAUUUGUUUGAUGAAGCCAGCAUCU SEQ ID NO:4 nCoV-S-5 ACUCUGACAUUUUAGUAGCAGCAGCAAGAUUAG SEQ ID NO:5 nCoV-S-6 AGCAGGAUCCACAAGAACAACAGCCCUUGA SEQ ID NO:6 nCoV-S-7 CAGAGACAUGUAUAGCAUGGAACCAAGUAA SEQ ID NO:7 hRluc-1 ACAAAGAUGAUUUUCUUUGGAAGGUUCAGC SEQ ID NO:8 hRluc-2 CGAAGAAGUUAUUCUCAAGCACCAUUUUCU SEQ ID NO:9
[0067] Synthesize Oligo DNA corr...
Embodiment 3
[0076] Plasmid transfection and dual luciferase reporter gene detection.
[0077] 293T cells were inoculated into a 24-well plate in a certain amount so that the confluence of the cells reached about 80% after 24 hours.
[0078] Using Lipofectamine 2000 liposome transfection reagent, the pXR004 plasmid with gRNA cloned and the pXR001 plasmid expressing Cas13d (the vector map of pXR001 is shown in Figure 4shown), psiCHECK2-nCoV-S plasmid was co-transfected with the above 293T cells at a ratio of 1:1:1, the psiCHECK2-nCoV-S plasmid was transfected alone as the negative control group, and the cells not transfected with any plasmid were used as the blank control . Three replicate wells (A / B / C) were set up for each transfection. The plasmid transfection groups are shown in Table 2.
[0079] Table 2. Plasmid transfection groups
[0080]
[0081]
[0082] 48 hours after the transfection of the plasmid, according to the method of using the dual-luciferase reporter gene dete...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com