DNAzymes for identifying pseudomonas aeruginosa as well as screening and detecting method and application
A Pseudomonas aeruginosa screening method technology, applied in the field of pathogenic bacteria detection and molecular biology, can solve the problem of detection of Pseudomonas aeruginosa with no DNAzyme, and achieve wide application value, strong specificity and effective detection Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
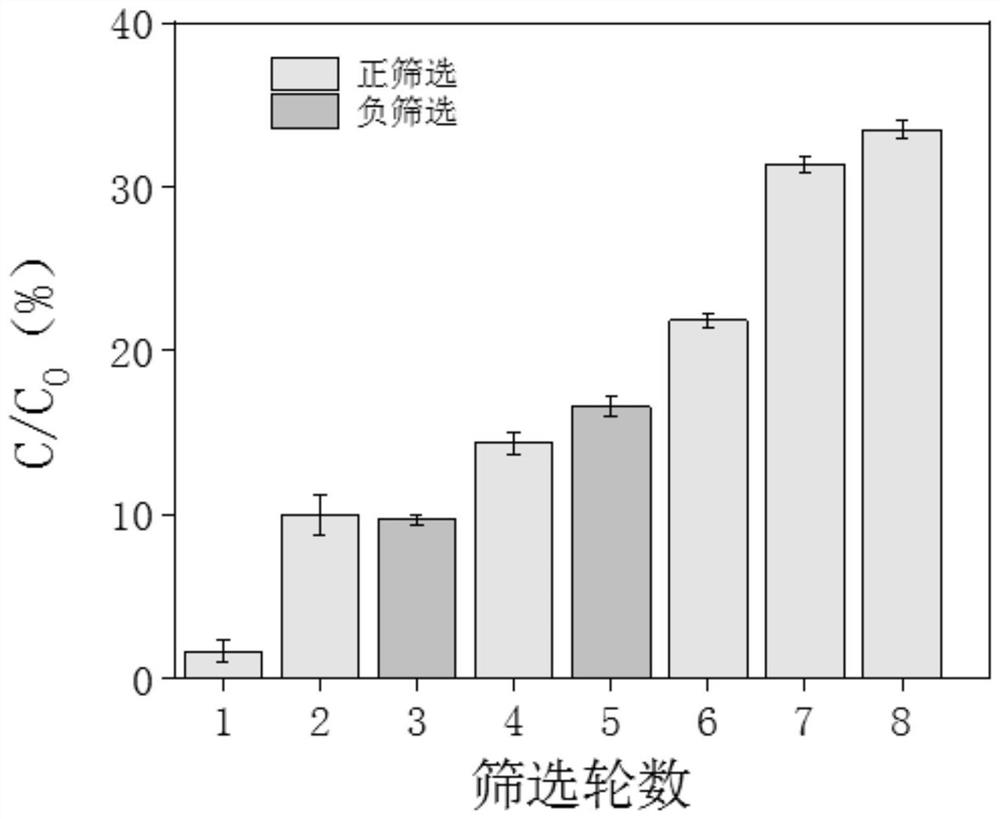
[0055] 1. Preparation before screening
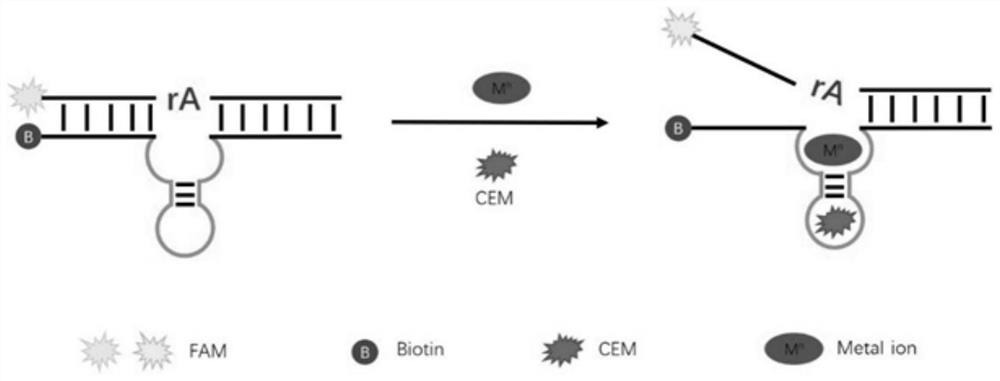
[0056] (1) Design of random library and PCR primers, the random library is a 75 nt sequence, and the 5' end is phosphorylated, the middle is a 35 nt random sequence, and the two sides are 20 nt primer sequences. Shanghai Sangon Bioengineering Co., Ltd. synthesized a DNA random library and related DNA sequences (5'-3') for in vitro screening.
[0057] Initial library: pGGACAAGAGGGGGATCTTGT-N 35 -GTTGTCAGCAGTCTGTCCAT
[0058] Primer 1: ATGGACAGACTGCTGACAAC
[0059] Primer 2: Biotin-TGCTGACAACTrAGGACAAGAGGGGGA
[0060] FAM-substrate: FAM-ATGGACAGACTGCTGACAACTrAGGACAAGAGGGGGA
[0061] (2) Pseudomonas aeruginosa was cultured with nutrient broth liquid medium, and the OD 600 nm The culture was stopped when it was close to 1, and the number of bacteria per milliliter was determined by the serial dilution coating method.
[0062] (3) Preparation of extracellular products: Divide 1 mL of all the culture solution into 1.5 mL sterile centrif...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com