Method and kit for detecting Helicobacter pylori drug resistance gene mutations
A Helicobacter pylori and kit technology, applied in biochemical equipment and methods, microbial measurement/testing, DNA/RNA fragments, etc., can solve problems such as time-consuming, complex and expensive equipment and processes, and limited detection throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
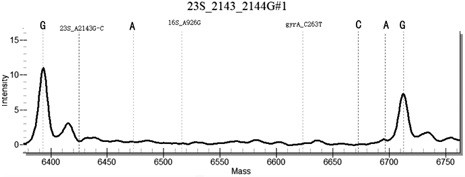
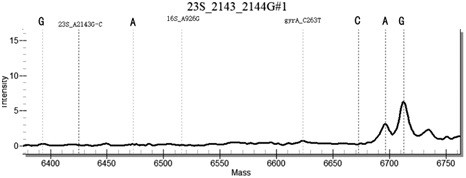
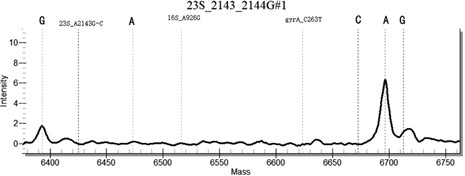
[0131] In the method for highly sensitive detection / or identification of drug resistance sites of Helicobacter pylori provided in this example, the samples to be tested include: some positive plasmids, positive samples with known drug resistance by sequencing, and negative sample controls. Including the following steps:
[0132] (1) For 43 common Helicobacter pylori drug resistance sites, design 43 specific site primers, select specific conserved sequences, design PCR primers, use a pair of PCR primers, with 10 at the 5' end of the primers The base (ACGTTGGATG) sequence, so that the total length reaches 29 bases or more, distinguishes the primer from the probe from the molecular weight. Design a single-base extension probe in the conserved sequence region in the amplification region, the length of the probe is 15-27 bases, and at the 3' end of the probe, it is allowed to extend a base determined by design as the genotype Specific sequence markers, with a total length of no mo...
Embodiment 2
[0153] Example 2 Specificity Determination
[0154] Using Escherichia coli, Staphylococcus aureus, Campylobacter jejuni, Candida albicans, Enterococcus faecium, Streptococcus pneumoniae, Salmonella paratyphi A and human genomic DNA nucleic acid as templates, multiplex PCR-time-of-flight mass spectrometry detection kit of the present invention As a result, no false positive results occurred, indicating that the kit of the present invention has good specificity.
Embodiment 3
[0155] Example 3 Clinical Sample Detection
[0156] Using the method for highly sensitive detection / or identification of Helicobacter pylori drug-resistant loci established in the present invention, the nucleic acid samples of 100 cases of Helicobacter pylori positive patients were detected, and the commercially available single-plex fluorescent quantitative PCR detection kit was used for verification.
[0157] The results show:
[0158] Among the 100 positive samples, both the single-plex fluorescent quantitative PCR and the multiplex PCR-time-of-flight mass spectrometry detection method of the present invention detected 100 positive samples, and the positive detection rate was 100%. Among them, 7 cases of tetracycline resistance, 3 cases of clarithromycin resistance, 2 cases of quinolone resistance, 18 cases of amoxicillin resistance, 15 cases of metronidazole resistance were identified, and the remaining samples were infected by sensitive strains.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com