CRISPR-Cas system for diagnosing spinal muscular atrophy and application thereof
A technology for spinal muscular atrophy and diagnostic reagent, applied in the field of CRISPR-Cas system for the diagnosis of spinal muscular atrophy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
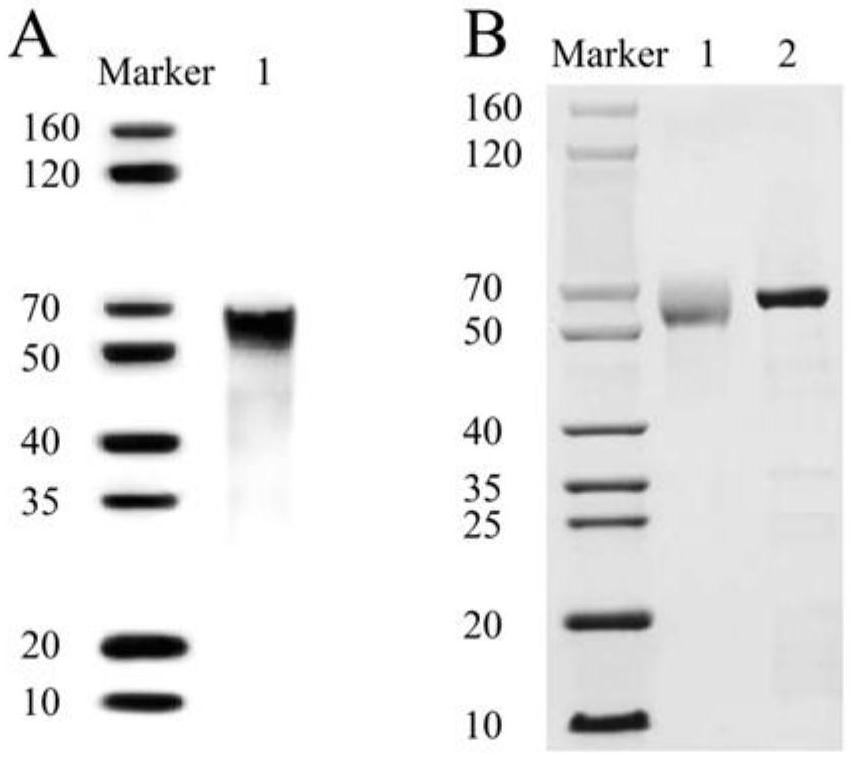
[0062] Cas14a1 protein expression and purification
[0063] The Cas14a1 expression vector was constructed and Cas14a1 protein was expressed in Escherichia coli, purified by affinity chromatography, and Cas14a1 protein was detected by western blotting.
[0064] 1) Cas14a1 protein expression
[0065] ①Transform the constructed plasmid containing the Cas14a1 gene into BL21(DE3) competent cells, evenly spread it on the LB plate containing 50 μg / mL kanamycin sulfate, and place it upside down in a 37°C incubator for overnight culture. Pick a single clone colony from the transformed plate and inoculate it into 4 mL of liquid LB medium containing 50 μg / mL kanamycin sulfate;
[0066] ②To be cultured to OD 600 0.5-0.8, add IPTG with a final concentration of 0.2mM to the LB culture medium, and then place them at 15°C and 37°C for 16 hours to induce expression;
[0067] ③ Centrifuge the induced culture medium at 12,000 rpm for 5 minutes, remove the supernatant, add PBS to resuspend the...
Embodiment 2
[0073] Detect the genotyped DNA and verify the method
[0074] Using normal human gDNA as a template, CRISPR / Cas14a1 cleavage detection was carried out after PCR amplification. By comparing the fluorescence increase values produced by different buffers, the optimal buffer was Cutsmart buffer of NEB (recipe: 50mM KAc, 20mM Tris-Ac, 10mM Mg (Ac)2, 100 μg / mL BSA (pH7.9@25°C)). In the subsequent detection process, we all choose Cutsmart buffer as the buffer.
[0075] 1. crRNA transcription
[0076] (1) Design and synthesize the corresponding DNA sequence of crRNA as shown in the above table, and use HiScribe in vitro TM T7High Yield RNA Synthesis Kit (NEW ENGLAND BioLabs, E2040s) transcription kit was used for transcription to obtain crRNA.
[0077] The transcription system is as follows:
[0078]
[0079] Incubate at 37°C for 16 hours.
[0080] (2) Remove possible residual DNA in the transcription system by DNaseI
[0081]
[0082] Incubate at 37°C for 30 minutes. ...
Embodiment 3
[0100] Comparing the impact of FQ-probe of different lengths
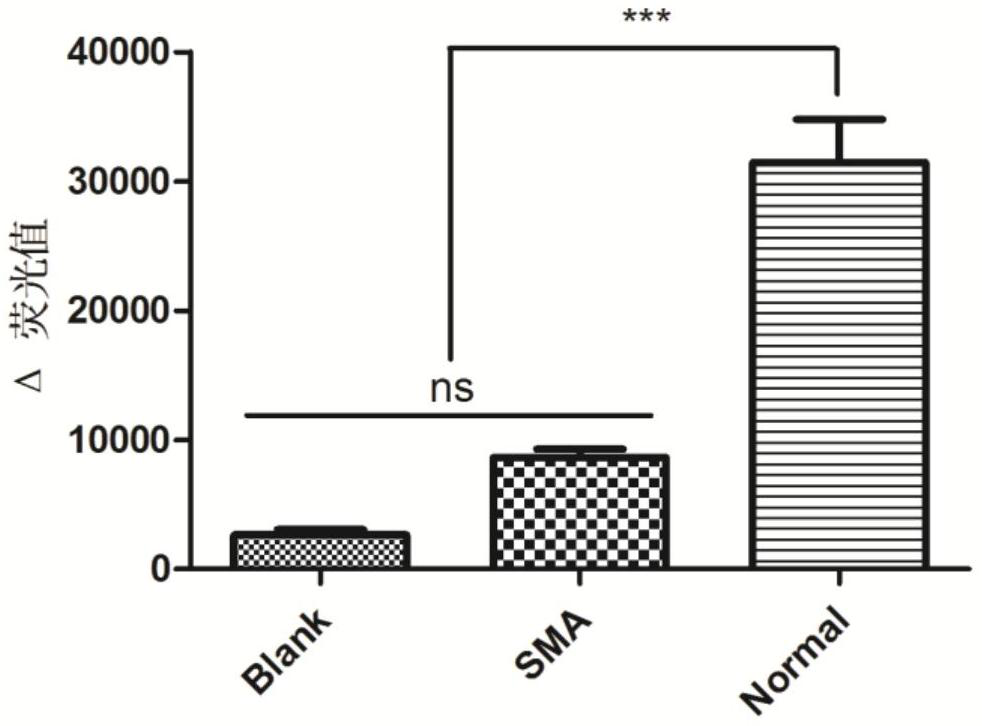
[0101] We constructed FQ-probes with a length of 12nt and 21nt, and used the normal human gDNA amplification product as a template to perform CRISPR / Cas14a1 cleavage fluorescence detection, and the specific steps were similar to Example 2. The results showed that the Δ fluorescence value produced by the 21nt FQ-probe was significantly higher than that produced by the 12nt FQ-probe ( Figure 4 ), so we use a 21nt length FQ-probe.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com