High-throughput pichia pastoris screening method based on droplet micro-fluidic chip
A microfluidic chip, Pichia pastoris technology, applied in the field of microbial biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 Droplet microfluidic high-throughput screening method for high-yield xylan of Pichia pastoris
[0031] 1. Construction and expression of Pichia pastoris strain with xylanase fusion green fluorescent protein
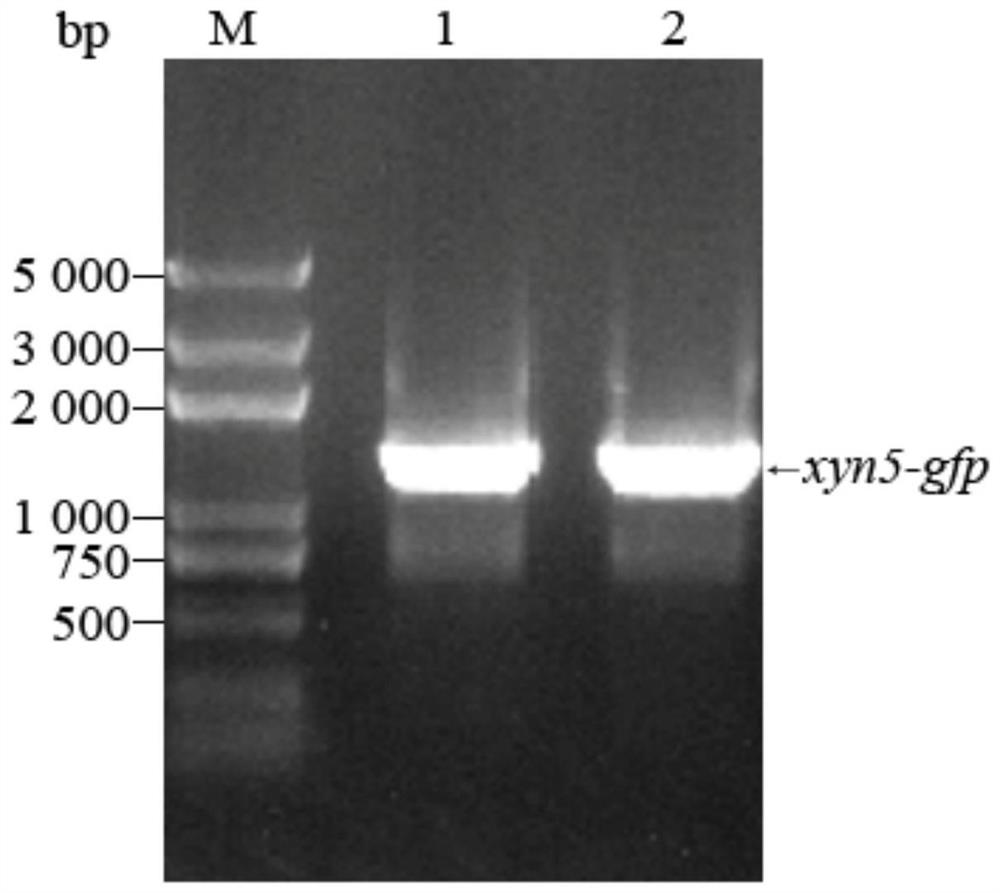
[0032] The fusion fragment of xylanase xyn5 gene and green fluorescent protein gfp gene was amplified by PCR, and cloned into the expression vector pPIC9K of Pichia pastoris to construct the plasmid pPIC9K-xyn5-gfp of xylanase fusion green fluorescent protein, using EcoRI It was verified by double enzyme digestion with NotⅠ. From Figure 2(a), two bands of xyn5-gfp fusion fragment and pPIC9K vector fragment can be seen; the plasmid was electrotransformed into Pichia pastoris GS115, and the genome was extracted for PCR amplification. To increase the integration efficiency, the results of agarose gel electrophoresis in Figure 2(b) show that the amplified band is consistent with the size of the exogenous gene fragment xyn5-gfp, and the recombinant Pichia past...
Embodiment 2
[0051] Droplet microfluidic high-throughput screening method for high-production cellulase from Pichia pastoris
[0052] Cellulase-producing Pichia strain GS115 / pPIC9K-cbh1 was constructed using the cbh1 gene derived from Penicilliuoxalicum, and the strain was treated according to steps 2-4 in Example 1, wherein the strain was mutagenized by ARTP to establish a mutant library, the mutagenesis time was 60s, and the strains in the Pichia mutant library were sonicated for 1min (the total power of the instrument was 950W, the power used was 4%, the ultrasound was 9.9s, and the stop was 9.9s, a total of 1min), and then embedded in OD 600 =0.3 for single droplet embedding and droplet culture, at 30°C, after 40 hours of incubation, detect the fluorescence signal intensity of the cellulase substrate in the droplet for microfluidic sorting of the droplet, and screen out the one with higher signal intensity or Several liquid droplets were subjected to shake-flask fermentation to screen ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com