Porcine epidemic diarrhea virus S gene complete sequence amplifying method and application thereof
A porcine epidemic diarrhea and full sequence technology is applied in the field of full sequence amplification of porcine epidemic diarrhea virus S gene, which can solve the problems of inability to amplify target fragments in hypervariable regions and heavy workload, so as to avoid inaccurate sequence information, Improve the success rate and reduce the effect of amplification failure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] A method for amplifying the full sequence of the S gene of porcine epidemic diarrhea virus, including the following steps:
[0034] Step 1. Extract viral RNA: Take an appropriate amount of porcine epidemic diarrhea virus-positive fecal material, add 5 times the volume of sterile PBS, and store at -20°C after homogenization; repeated freezing and thawing 3 times, using Trizol reagent method to extract the virus RNA;
[0035] The specific method of the Trizol reagent method is: a. Take 1 ml of the tissue suspension and centrifuge at 5000 rpm for 10 min at 4°C to obtain the first supernatant; b. Take 300 μL of the first supernatant. Add 500μL of Trizol to the RNase-free sterilized 1.5ml EP tube, mix well, and let stand at room temperature for 10min; c. Add 500μL of chloroform, mix well, let stand at room temperature for 10min, at 4℃, Centrifuge at 12000 rpm for 10 min to obtain the second supernatant; d. Take 500 μL of the second supernatant into a new 1.5 mL EP tube, add 1.0 ...
Embodiment 2
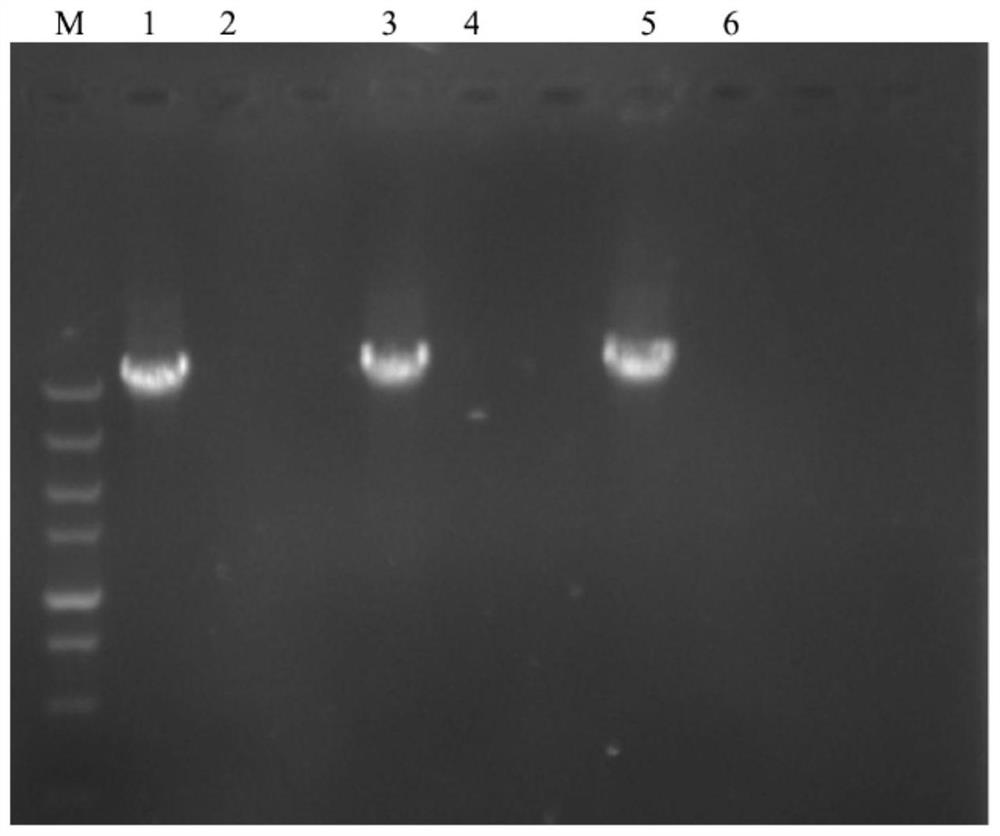
[0044] A method for amplifying the full sequence of the S gene of porcine epidemic diarrhea virus. The disease material used in step 1 is the porcine epidemic diarrhea virus-positive small intestine disease material. The remaining steps are the same as in Example 1. In the gene fragment detection and sequencing steps, the electrophoresis results Such as figure 1 The lane 3 in is shown, and the sequencing result is shown in SEQ ID NO:5. The PEDV strain contained in the disease material sample was named LY-201803.
Embodiment 3
[0046] A method for amplifying the full sequence of the S gene of porcine epidemic diarrhea virus. The disease material used in step 1 is the porcine epidemic diarrhea virus-positive small intestine disease material. The remaining steps are the same as in Example 1. In the gene fragment detection and sequencing steps, the electrophoresis results Such as figure 1 Lane 5 in is shown, and the sequencing result is shown in SEQ ID NO:6. The PEDV strain contained in the disease material sample was named LY-201805.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com