Single cell transcriptome sequencing method based on third-generation sequencing
A single-cell, transcriptome technology, applied in the field of single-cell sequencing, can solve problems such as material inaccessibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
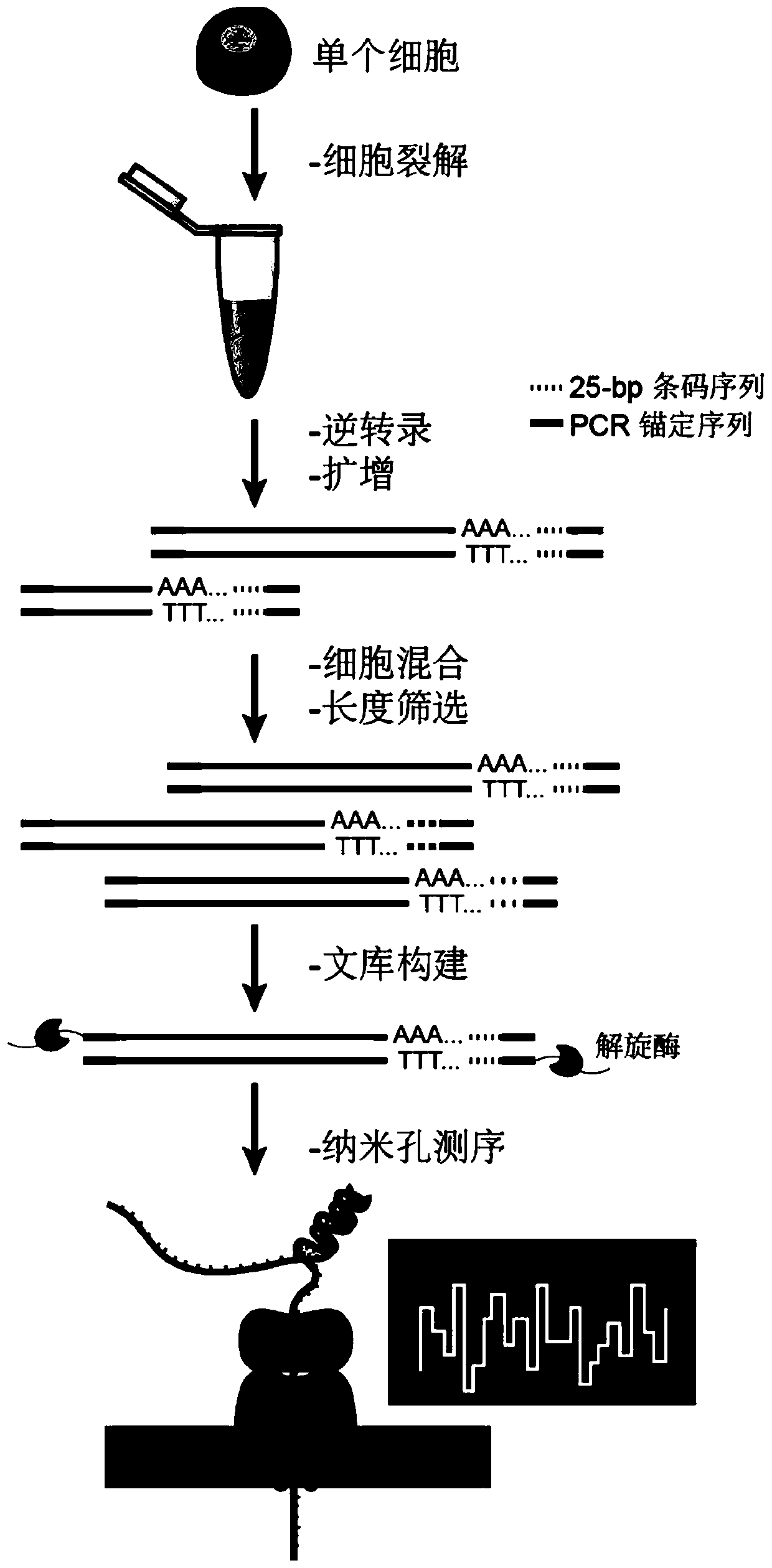
[0057] Example 1 Principle of single-cell transcriptome sequencing
[0058] The flow chart of single-cell transcriptome sequencing based on nanopore single-cell sequencing of the present invention is as follows figure 1 Shown:
[0059] The single-cell lysate is co-incubated with the reverse transcription system, and the reverse transcription primer is combined with the polyA tail of the single-cell mRNA through poly dT, and the barcode sequence is introduced into the single-cell mRNA, and the mRNA is reverse-transcribed into the first strand under the action of reverse transcriptase cDNA, the barcode sequence realizes the specific labeling of the first-strand cDNA; under the action of terminal transferase, the template-switching primer combines with the first-strand cDNA and extends to obtain double-stranded DNA;
[0060] Using the anchor sequence to perform PCR amplification to obtain the amplified product; using magnetic beads to purify the amplified product, removing the a...
Embodiment 2
[0062] Example 2 Single Cell RNA Reverse Transcription
[0063] (1) Single cell lysis
[0064] Isolate single mouse embryonic stem cells (mESCs), transfer each cell to an independent reaction tube, and add a certain amount of single cell lysate to prepare the lysis system shown in Table 1, shake and mix and incubate at 72°C for 3 minutes to make The cells are fully lysed to release RNA, and the samples are immediately transferred to ice;
[0065] Table 1 Single cell lysis system
[0066] Element Volume (μL) 10% Triton X-100 0.095 RNase inhibitor (40U / μL) 0.05 Reverse transcription primer (5μM) 0.3 dNTPs (10mM) 0.5 Enzyme-free water 1.555
[0067] (2) In vitro reverse transcription reaction
[0068] Add the reverse transcription reaction solution shown in Table 2 directly to the single-cell lysed sample obtained in step (1). °C for 10 min to obtain cDNA complementary to the RNA, and the cDNA carries a barcode sequence (Barcode)....
Embodiment 3
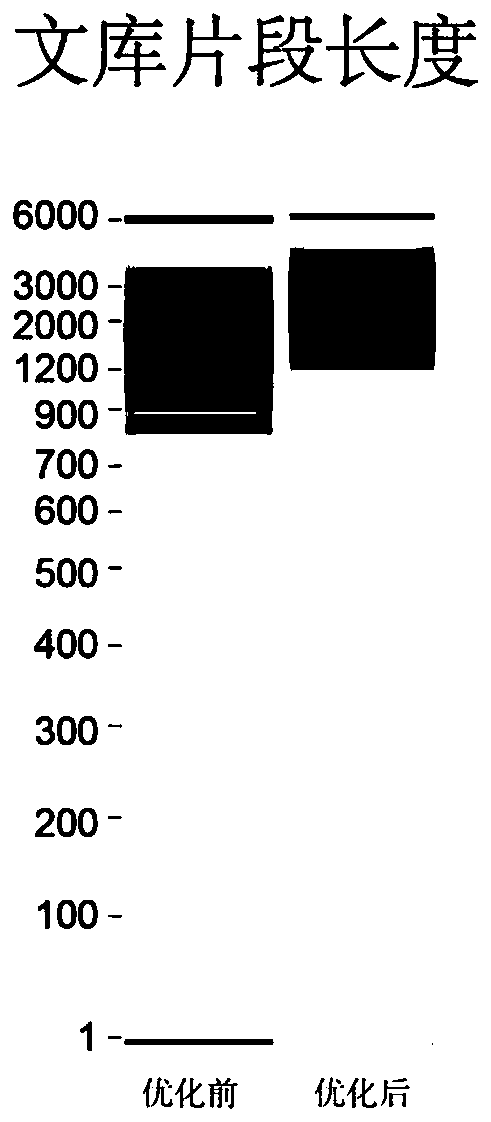
[0071] Example 3DNA amplification and amplification product purification
[0072] (1) PCR amplification
[0073] Perform PCR amplification reaction on the cDNA obtained in Example 2. The system is shown in Table 3. The conditions are 95°C pre-denaturation for 3 minutes; 98°C denaturation for 20s, 65°C annealing for 30s, 72°C extension for 5min, 3-6 cycles; 98°C Denaturation at ℃ for 20s, annealing at 67℃ for 15s, extension at 72℃ for 5min, 10-20 cycles;
[0074] Table 3 PCR amplification system
[0075] Element Volume (μL) KAPA HiFi HotStart ReadyMix (2×) 6.25 Anchor primer (10 μM) 0.25 cDNA template 5.4 Enzyme-free water 1
[0076] (2) Product purification
[0077] Add SPRI magnetic beads to purify the amplified product. The magnetic beads are 0.4 times the volume of the reaction solution. During the purification process, the magnetic beads and the product are mixed first, incubated at room temperature for 5 minutes, and then placed ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com