Application of PGAM5 as diagnostic marker and therapeutic target for oligospermia and asthenozoospermia
A technology for asthenozoospermia and oligospermia, applied in the field of oligospermia diagnostic markers, can solve the problems of little understanding, and achieve the effect of high accuracy and diagnostic value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] 1. Detection method
[0028] In this example, a method for detecting oligospermia markers based on sperm proteomics is provided. The samples used in the following examples are all from the Reproductive Hospital Affiliated to Shandong University. The forward movement of sperm in samples from patients with oligoasthenospermia is 7.8 %-17.3, the sperm concentration is between 3.8-14.8 million / mL, the detection method includes the following steps:
[0029] (1) Extract the total protein of sperm cells: wash the sperm sample with DPBS, add RIPA lysis buffer and ultrasonic for 1 to 2 minutes, incubate on ice for 30 minutes to lyse, centrifuge at 14,000g for 20 minutes, take the supernatant protein liquid, use the Bradford method to determine the liquid Medium protein concentration.
[0030] (2) Enzymatically digest the protein using the method based on ultrafiltration assisted sample preparation (FASP): add 300μg protein solution to a 10KD ultrafiltration tube, shake at 600RPM for 1...
Embodiment 2
[0049] Example 2 Clinical test verification
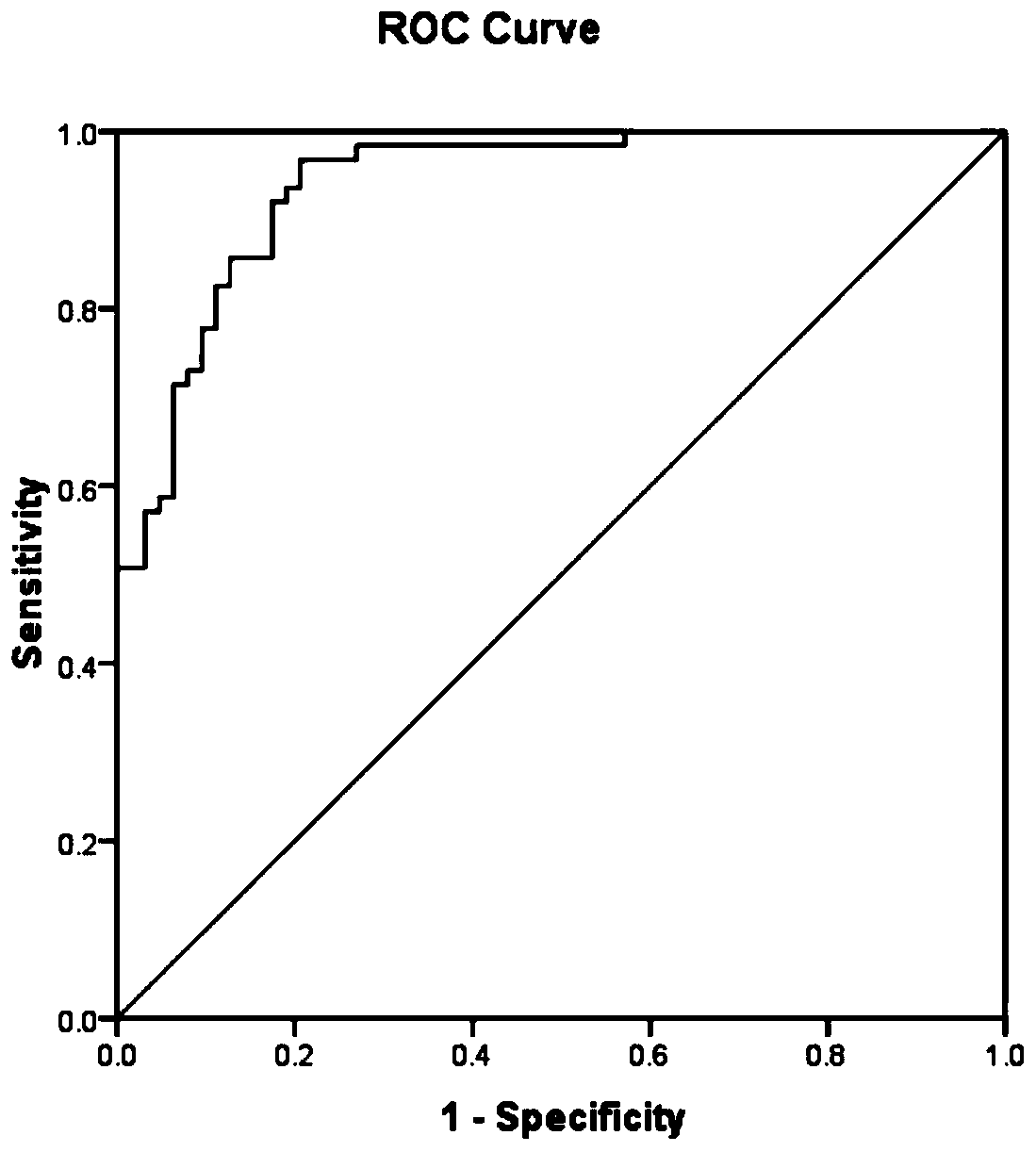
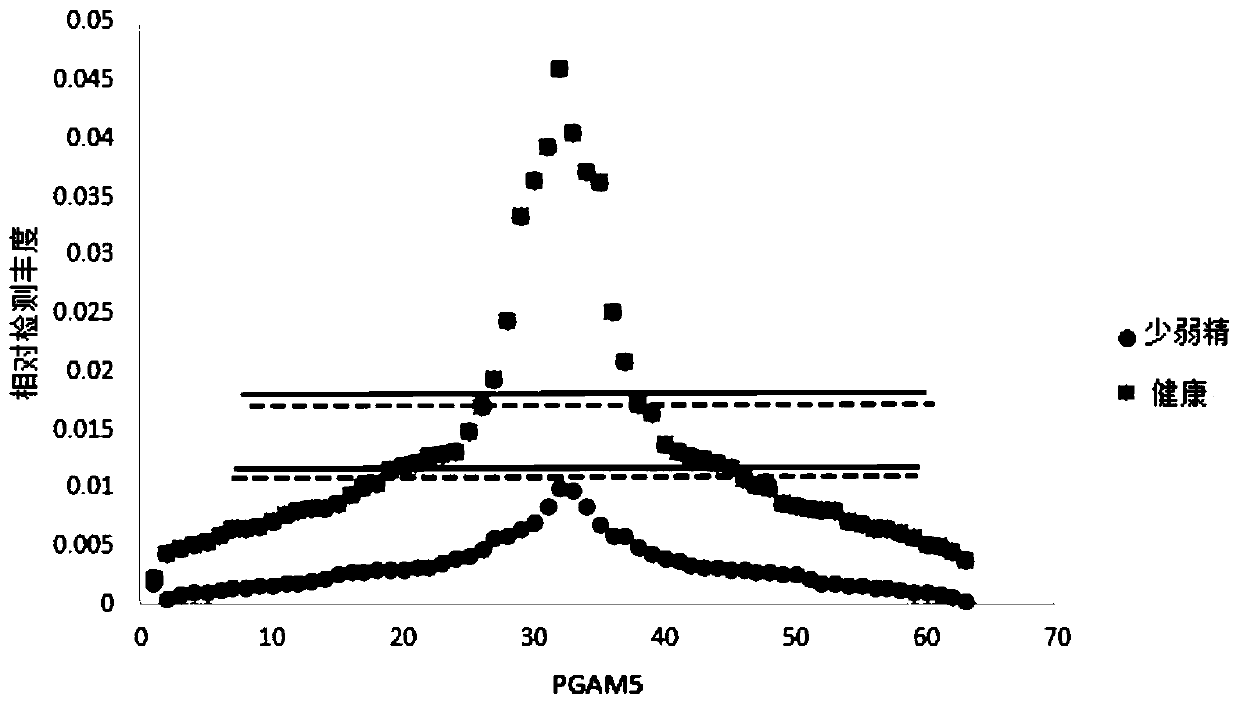
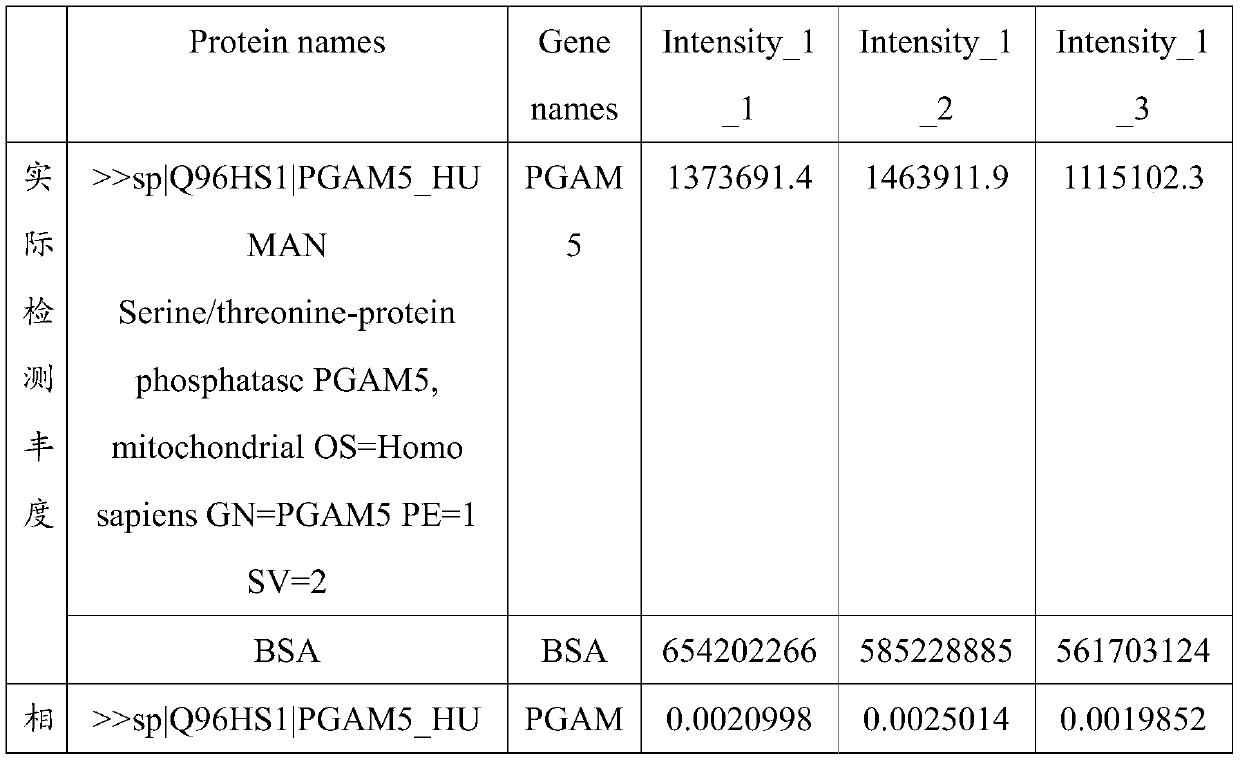
[0050] According to the method of Example 1, the collected sperm of 10 groups of oligoasthenospermia patients were firstly subjected to protein extraction and nanoflow liquid chromatography-tandem mass spectrometry analysis, and each set of data was repeated three times, and then the mass spectrometry data was analyzed. Table 1 shows the results of the analysis. 10 people were detected with oligoasthenospermia using the above method. The accuracy of this set of data is as high as 100%.
[0051] Table 1
[0052]
[0053]
[0054] Continued
[0055]
[0056] Continued
[0057]
[0058] Continued
[0059]
[0060]
[0061] Continued
[0062]
[0063] Continued
[0064]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com